Docking method for proteins, water molecules and ligands
A water molecule and protein technology, applied in molecular design, multi-objective optimization, design optimization/simulation, etc., can solve problems such as high computational complexity, easy to fall into local minimum, etc., to improve search performance, avoid repeated parameter debugging, The effect of increasing quantity and variety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
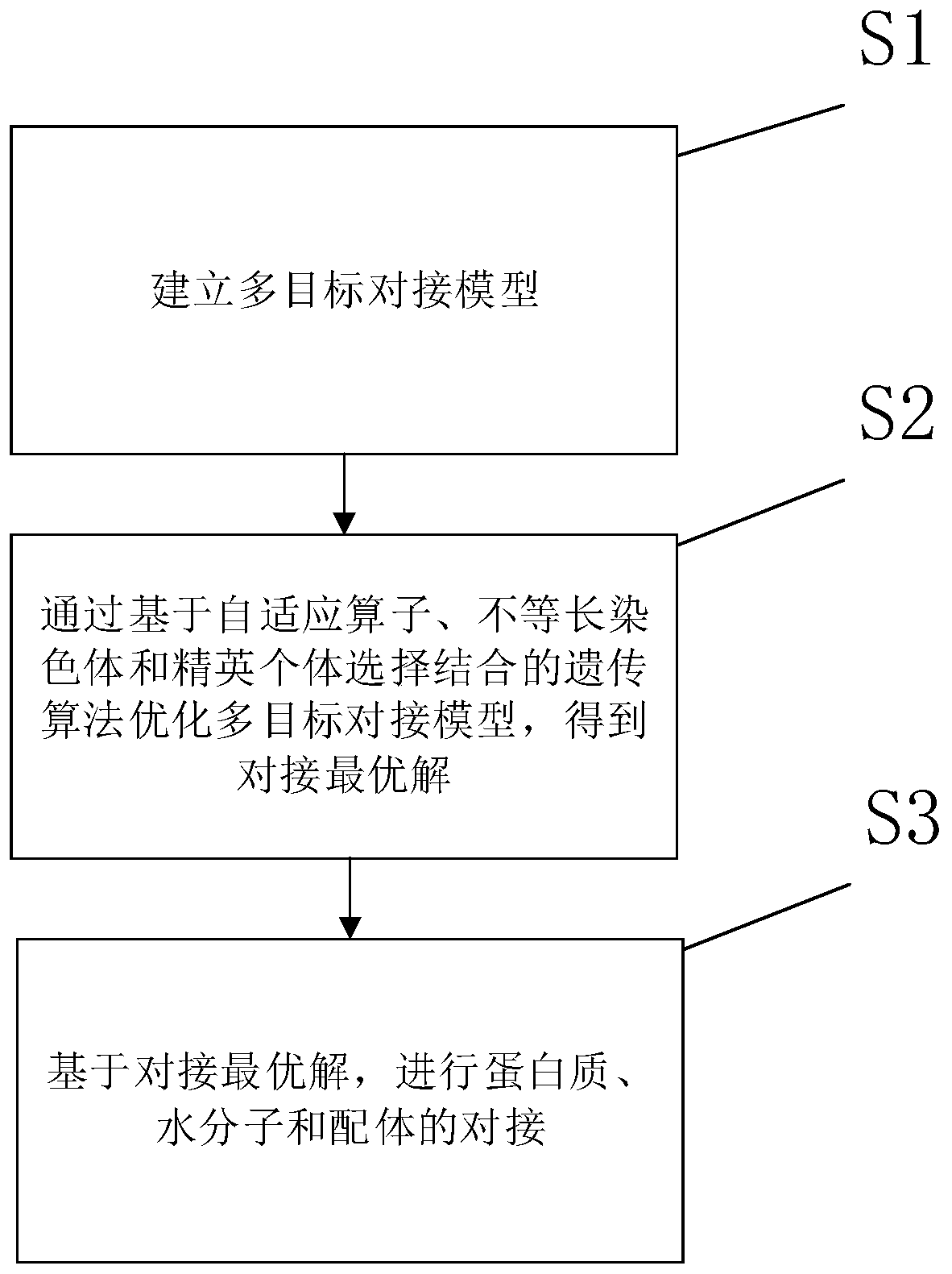
[0059] This example provides a docking method suitable for proteins, water molecules and ligands with an uncertain number of molecules, which can avoid falling into a local optimum and has low computational complexity, such as figure 1 shown, including the following steps:
[0060] Step S1: Establish a multi-target docking model;
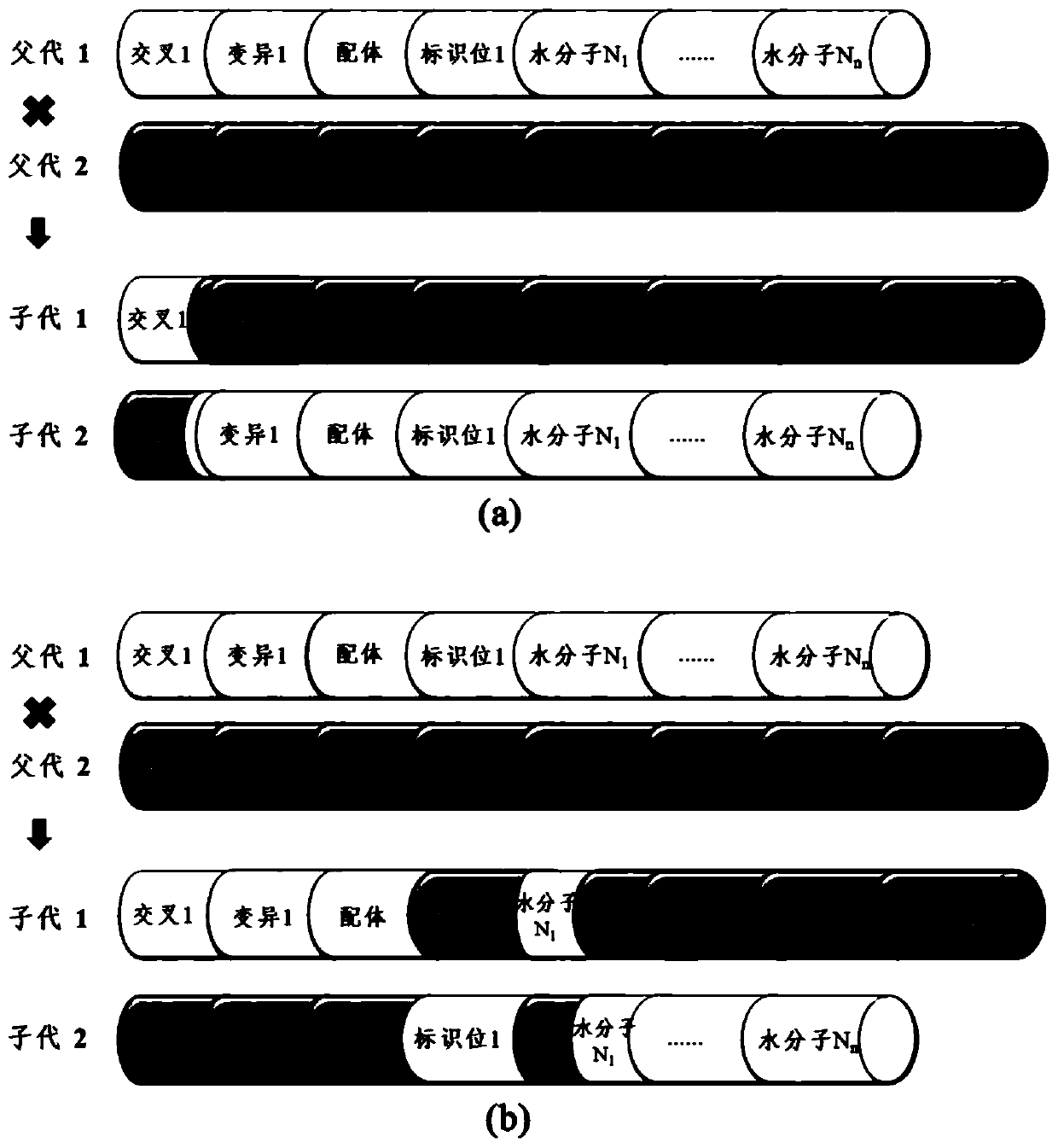
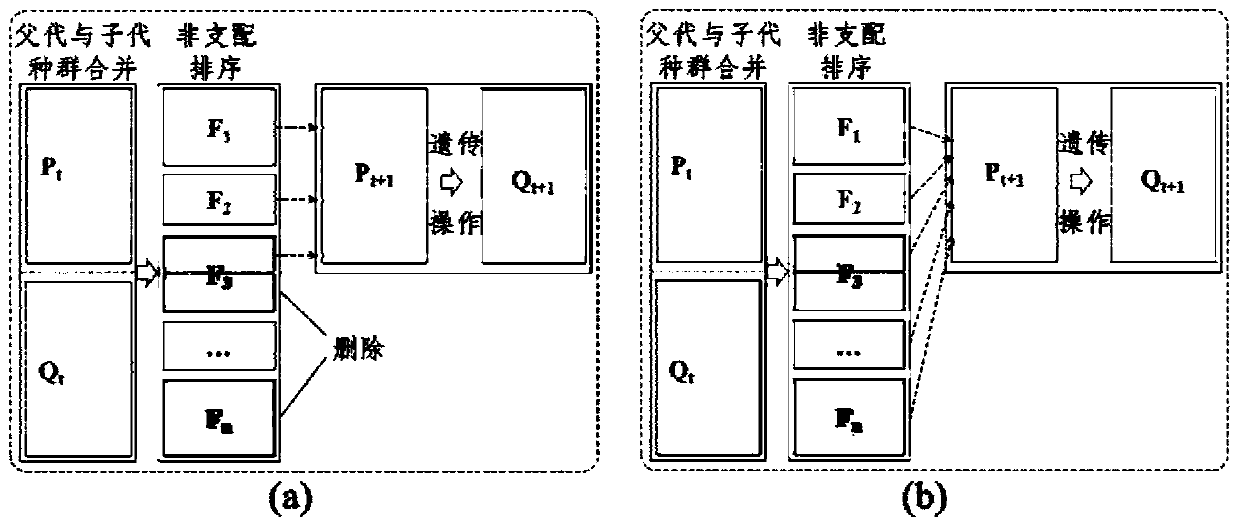
[0061] Step S2: optimize the multi-objective docking model through the genetic algorithm based on the combination of adaptive operators, unequal length chromosomes and elite individual selection, and obtain the optimal docking solution;
[0062] Step S3: Based on the optimal docking solution, the docking of proteins, water molecules and ligands is carried out.
[0063] in particular:
[0064] 1) Adaptive operator
[0065] For the two important control parameters of the genetic algorithm, the crossover probability and the mutation probability, let them participate in the entire evolution process as optimal design variables, so that the changes of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com