Sequence and application of a nanobody H6 that specifically binds to cancer cell protein b7-h4
A technology of B7-H4 and nanobody, which is applied in the sequence and application field of nanobody H6, can solve the problems of high immunogenicity, low tumor targeting, and unsuitability, and achieve low immunogenicity, affinity and stability The effect of high performance and simple structure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
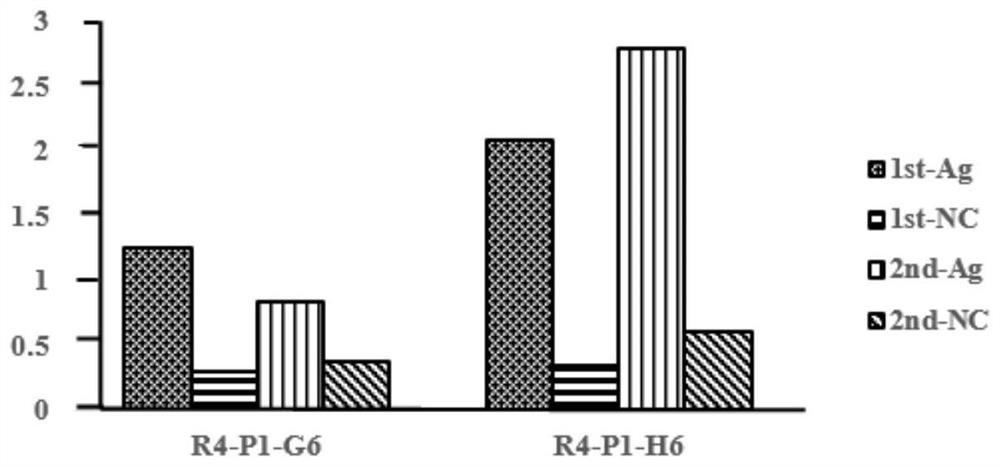
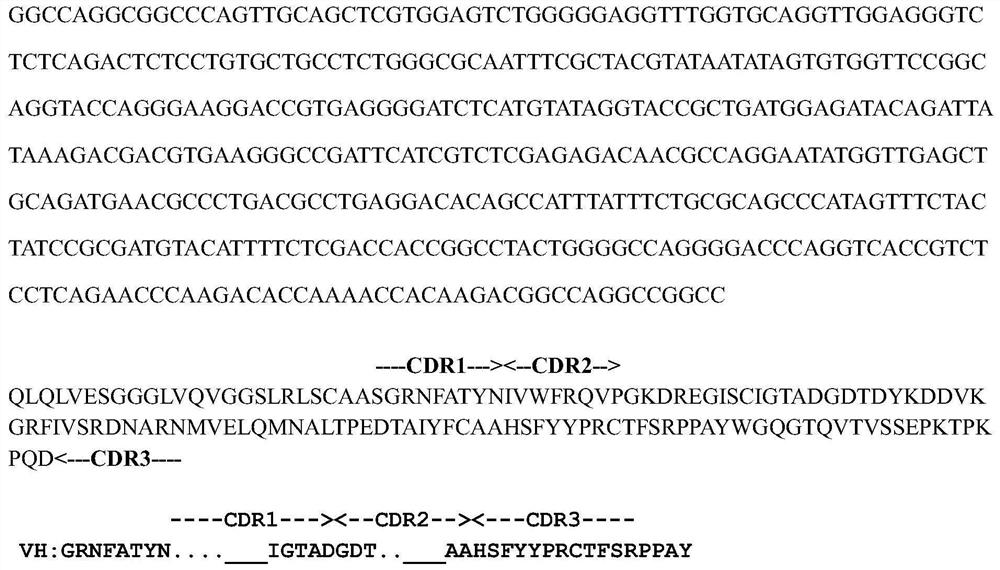
[0062] Example 1 Screening of Nanobodies Binding to Cancer Cell-Specific Protein B7-H4
[0063] Phage display antibody library technology mainly displays antibody molecules on the surface of bacteriophages, uses target antigen molecules to screen phages expressing specific antibody molecules, and expresses the antibodies and subsequent functions through genetic engineering methods. identification to obtain functional antibody molecules. The present invention adopts a non-immunized alpaca phage library (Lama-VHH Phage Display Library, constructed by collecting 20 non-immunized alpaca lymphocytes and one alpaca spleen cell). Total RNA from mixed cells was extracted and reverse transcribed into cDNA. PCR amplification of VHH segment gene sequence.
[0064] Phage display antibody library vector construction: The library was constructed with the pComb3XSS vector, and the pComb3XSS was digested into two large fragments of 1672bp (SS stuffer) and 3301bp (the target fragment of the ...
Embodiment 2
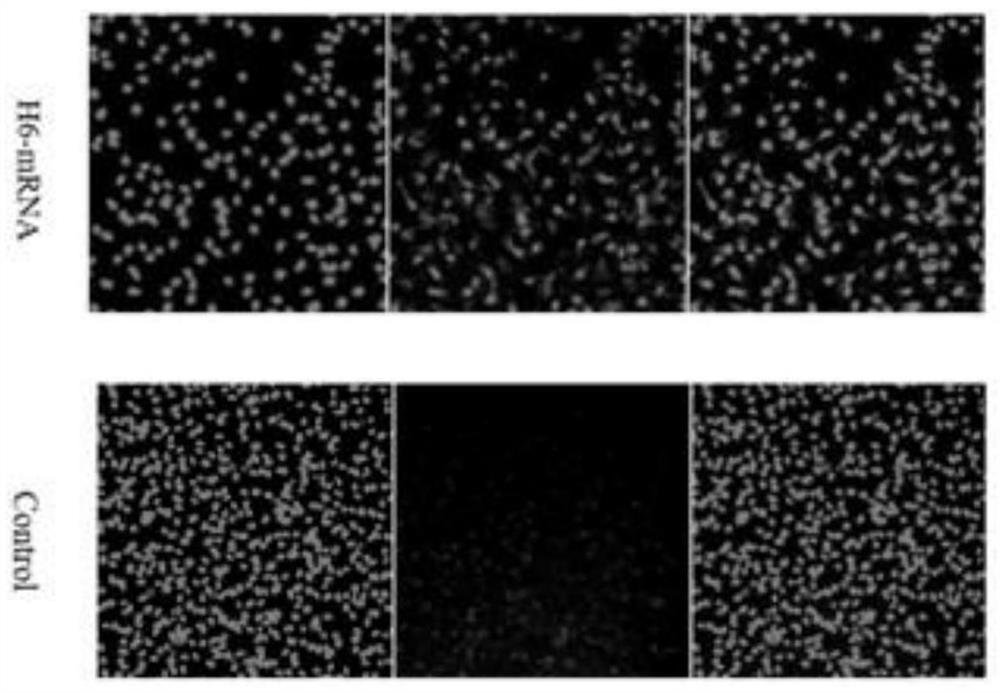
[0090] Example 2 Cellular Expression of Nanobodies
[0091] 1) In vitro synthesis of B7-H4 nanobody H6 mRNA
[0092] The leader peptide sequence IL-2 was added before the H6 coding DNA sequence obtained from the screening, and the His tag sequence was added after the sequence. These two sequences were synthesized by Shanghai Shenggong as PCR amplification primers, upstream primer TTGGACCCTCGTACAGAAGCTAATACG (27bp), downstream primer T 120 CTTCCTACTCAGGCTTTATTCAAAGACCA (149bp). The three segments were synthesized by PCR. The PCR product was transcribed in vitro by the in vitro transcription kit, and 3'-O-Me-m7G(5')ppp(5')GRNA cap analog was added during the in vitro transcription process. After the reaction, Turbo DNase (from MEGAscript T7 kit) was added and incubated on a thermal cycler at 37°C for 15 min. Add 10X antarctic phosphatase buffer and Antarctic phosphatase and purify with Monarch RNAclearupkit.
[0093] 2) Cell culture and immunofluorescence detection of the ex...
Embodiment 3
[0096] Example 3 CCK-8 method to detect the inhibitory effect of Nanobody H6 on cancer cell proliferation
[0097] The cell suspension transfected with H6 mRNA was placed in a 96-well plate (100 μL / well), pre-incubated for 24 h, and a blank group and a transfection reagent control group were set; 10 μL of CCK-8 solution was added to each well and shaken gently. , incubate for 1 h in a 37°C incubator; measure the absorbance at 450 nm with a microplate reader. Calculate percent cell inhibition. See Figure 8 .
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap