Base balance-based molecular marker method for amplicon
A technology of molecular labeling and base balance, which is applied in the field of molecular biology and can solve problems such as base imbalance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0030] 341F forward primer containing molecular markers:
[0031]
[0032]
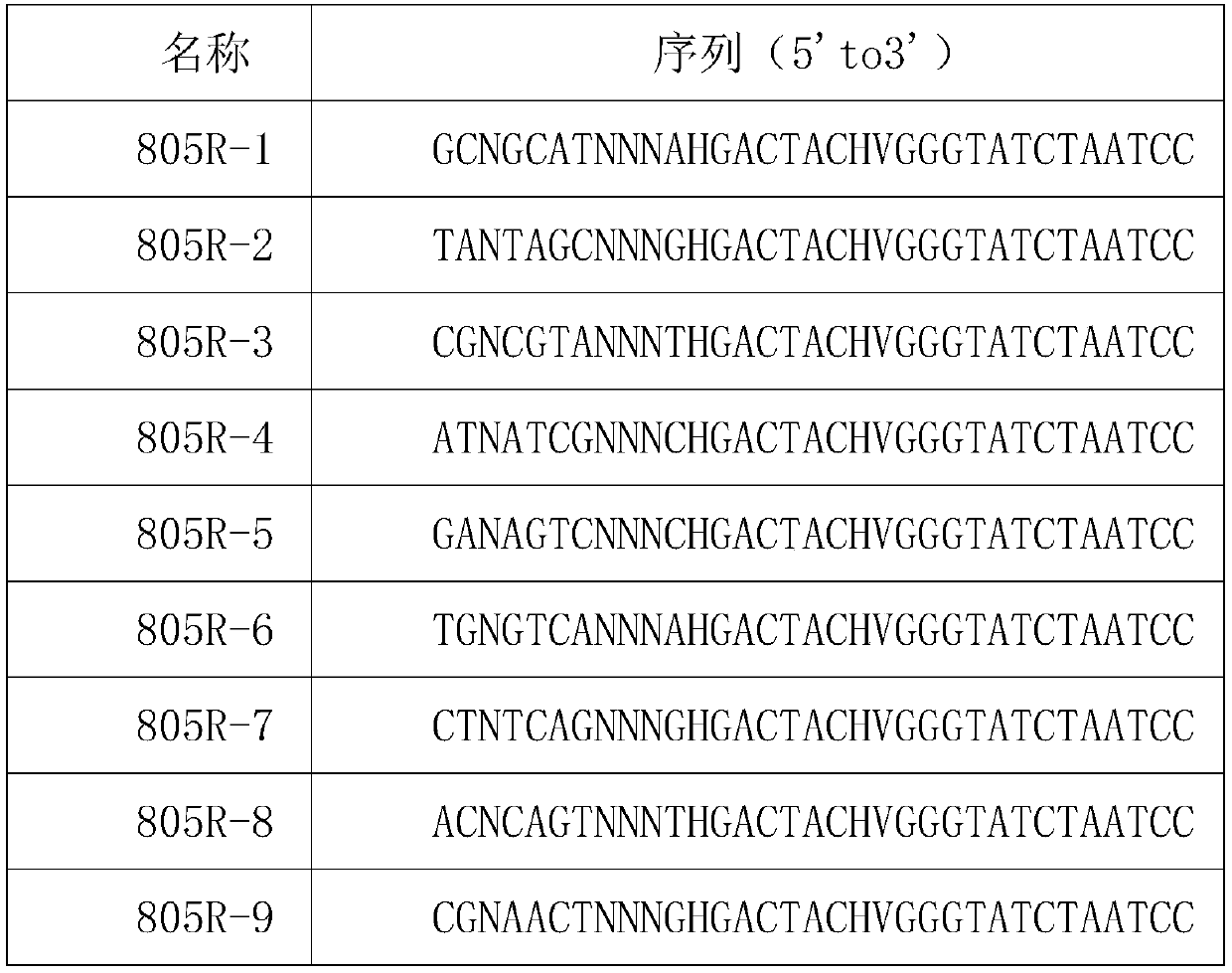
[0033] 805R reverse primer containing molecular markers:
[0034] name sequence(5'to3') 805R-1 GCNGCATNNNAHGACTACHVGGGTATCTAATCC 805R-2 TANTAGCNNNGHGACTACHVGGGTATCTAATCC 805R-3 CGNCGTANNNTHGACTACHVGGGTATCTAATCC 805R-4 ATNATCGNNNCHGACTACHVGGGTATCTAATCC 805R-5 GANAGTCNNNNCHGACTACHVGGGTATCTAATCC 805R-6 TGNGTCANNNNAHGACTACHVGGGTATCTAATCC 805R-7 CTNTCAGNNNGHGACTACHVGGGTATCTAATCC 805R-8 ACNCAGTNNNTHGACTACHVGGGTATCTAATCC 805R-9 CGNAACTNNNNGHGACTACHVGGGTATCTAATCC 805R-10 ATNGGANNNNTHGACTACHVGGGTATCTAATCC 805R-11 GCNTTGANNNCHGACTACHVGGGTATCTAATCC 805R-12 TANCCTGNNNAHGACTACHVGGGTATCTAATCC
[0035] 519F forward primer containing molecular markers:
[0036]
[0037]
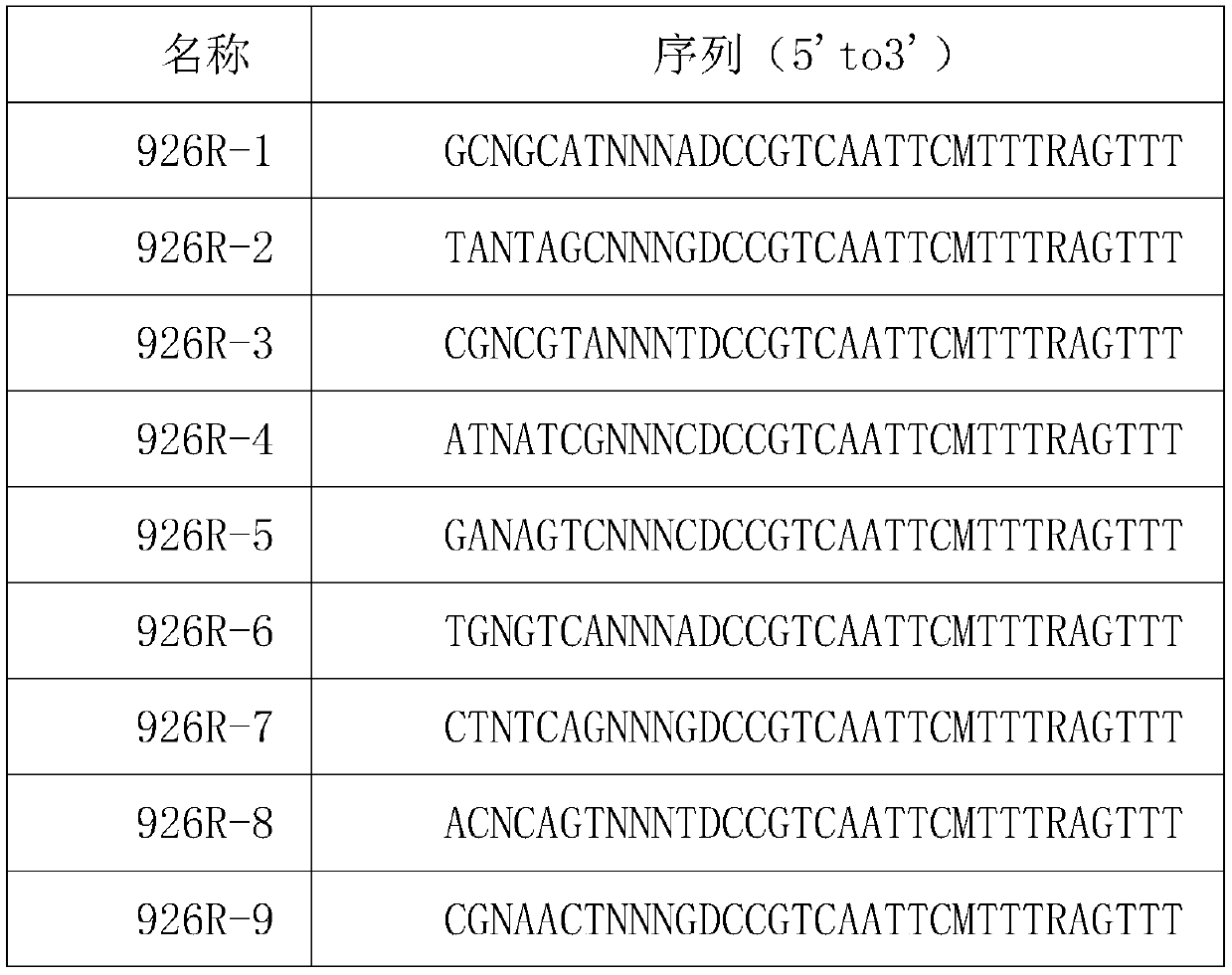
[0038] 926R reverse primer containing molecular markers:
[0039] name sequence(5'to3') 926R-1 GCNGCATNNNADCCGTCAATTCMTTTRAGTTTT 9...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com