Signal amplification magnetic bead technology system and application of nucleic acid detection based on CRISPR technology
A signal amplification and magnetic bead technology, applied in the field of signal amplification magnetic bead technology system, can solve problems such as missed detection and false negatives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
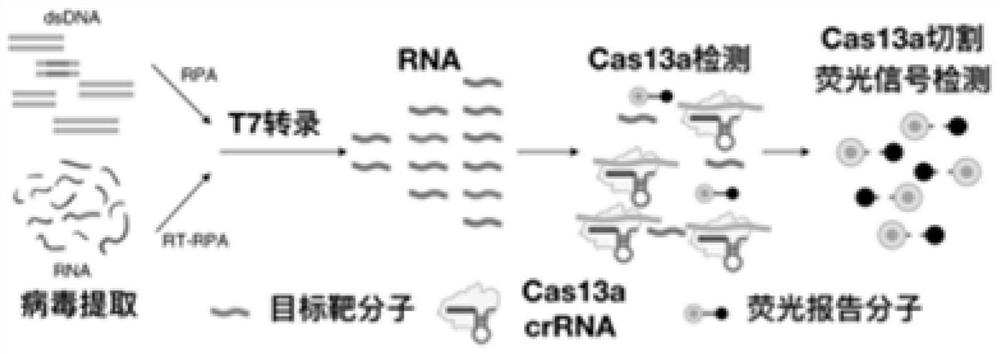
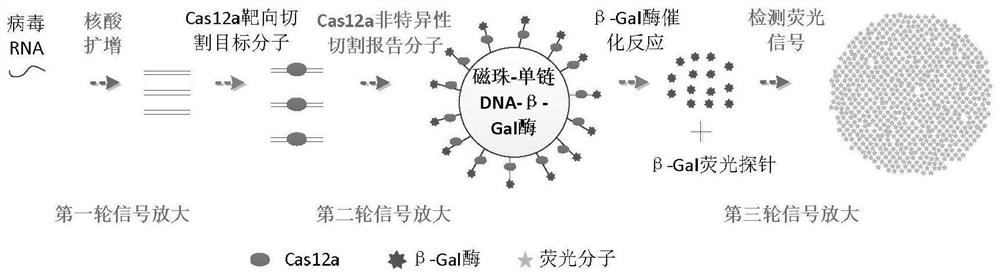
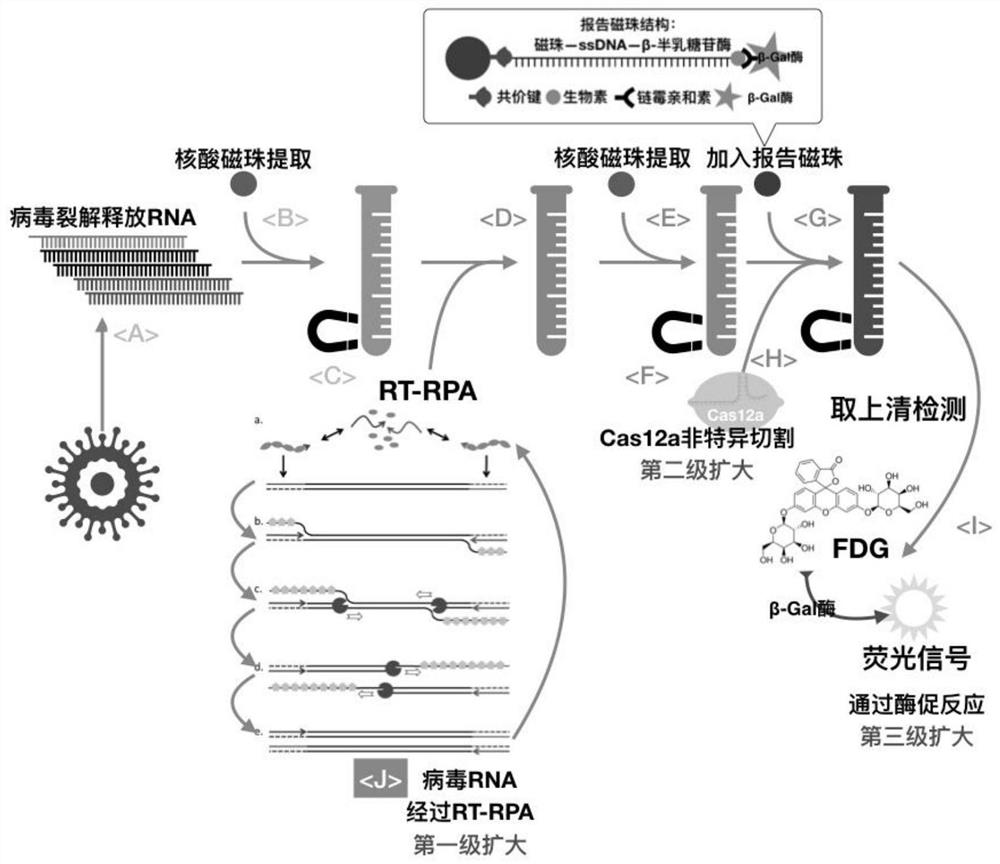
Image
Examples
Embodiment 1
[0063] Taking the coronavirus RNA detection method constructed by the Cas12a-based signal amplification magnetic bead technology system as an example to illustrate the specific technical scheme:
[0064] 1. Test sample: fake virus (reference substance)
[0065] Product name: COVID-19-pseudovirus (2019-nCOV pseudovirus)
[0066] Product number: 11900ES08 (Shanghai Yisheng Biotechnology Co., Ltd.)
[0067] Specification: 5×1mL
[0068] Target value = 1×10 8 copies / mL
[0069] 2. The sequence to be detected:
[0070] GGTTATGGCTGTAGTTGTGATCAACTCCGCGAACCCATGCTTCAGTCAGCTGATGCACAATCGTTTTTAAACGGGTTTGCGGTGTAAGTGCAGCCCGTCTTACACCGTGCGGCACAGGCACTAGTACTGATGTCGTATACAGGGCTTTTGACATC
[0071] 3. Isothermal amplification primer synthesis
[0072] RPA-F primer: GGTTATGGCTGTAGTTGTGATCAACTCCGC
[0073] RPA-R primer: GATGTCAAAAAGCCCTGTATACGACATCAGTAC
[0074] 4. Design and synthesis of crRNA
[0075] Design and synthesize crRNA sequence: AGACGGGCTGCACTTACACCG,
[0076] 5. Design and prepa...
Embodiment 2
[0128] Taking the Salmonella DNA detection method constructed by the Cas13a-based signal amplification magnetic bead technology system as an example to illustrate the specific technical scheme:
[0129] 1. Test sample: Salmonella standard plasmid (reference substance)
[0130]Salmonella standard plasmid preparation process: the Salmonella inv A gene (GenBank: KJ718885.1) was selected as a specific detection fragment, and the 287bp inv A gene fragment was connected to the pMD19-T vector to construct a standard plasmid.
[0131] Specification: 10×1mL
[0132] target value = 5 x 10 10 copies / mL
[0133] 2. The sequence to be detected:
[0134] TGTGAAATTATCGCCACGTTCGGGCAATTCGGTATTGACGATAGCCTGGCGGTGGGTTTTGTTGTCTTCTCTATTGTCACCGTGGTCCAGTTTATCGTTATTACCAAAGGTTCAGAACGCGTCGCGGAAGTCGCGGCCCGATTTTCTCTGGATGGTATGCCCGGTAAACAGATGAGTATTGATGCCGATTTGAAGGCCGGTATTATTGATGCGGATGCCGCGCGCGAACGGCGAAGCGTACTGGAAAGAGAAAGCCAGCTTTACGGTTCCTTTGACGGTGCGATGAA
[0135] 3. Design and synthesis of isothermal amp...
Embodiment 3
[0190] Taking the detection method of whether Schistosoma mansoni is contained in the water sample in the epidemic area constructed by the signal amplification magnetic bead technology system based on Cas12a as an example to illustrate the specific technical scheme:
[0191] 1. The samples to be tested are: water samples confirmed to be slightly polluted by Schistosoma mansoni (A), water samples confirmed to be moderately polluted by Schistosoma mansoni (B), water samples confirmed not to contain Schistosoma mansoni ( C), samples of Schistosoma mansoni (D)
[0192] 2. DNA extraction of samples to be tested: four DNA samples of A, B, C, and D were extracted;
[0193] 3. The sequence to be detected
[0194] CCTTCGGGCATTGCTGAGTGTGGTCGGTTTGTTACTAGCTTCGGCTGGTCGGCTGATGGCTTGGTTTTGTCACGTCGGCGGTTGCGTGTGTGGTTTGCATTGGGCCAATAGTCTGTGGTGTAGTGGTAGACGATCCACCTGACCCGTCTTGAAACACGGACCAAAGGAGTTTAACATGTGCGCGAGTCATTGGGGTGTAGTAGTAGTCAGC
[0195] 4. Design and synthesis of isothermal amplification p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com