Methods for identifying biological material by microscopy
A technology of biomaterials, learning methods, applied in the domain of discrete objects with optional quantification of biological origin for the identification of opinions, able to solve the problems of low counts, false negative results, loss, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
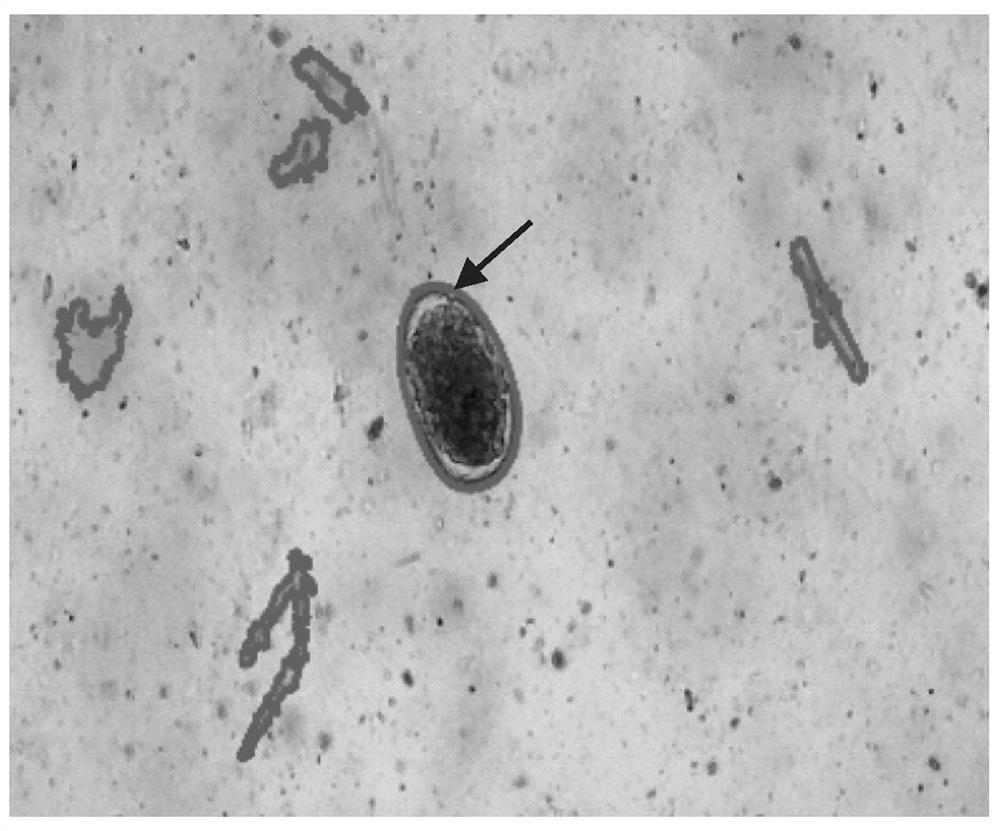
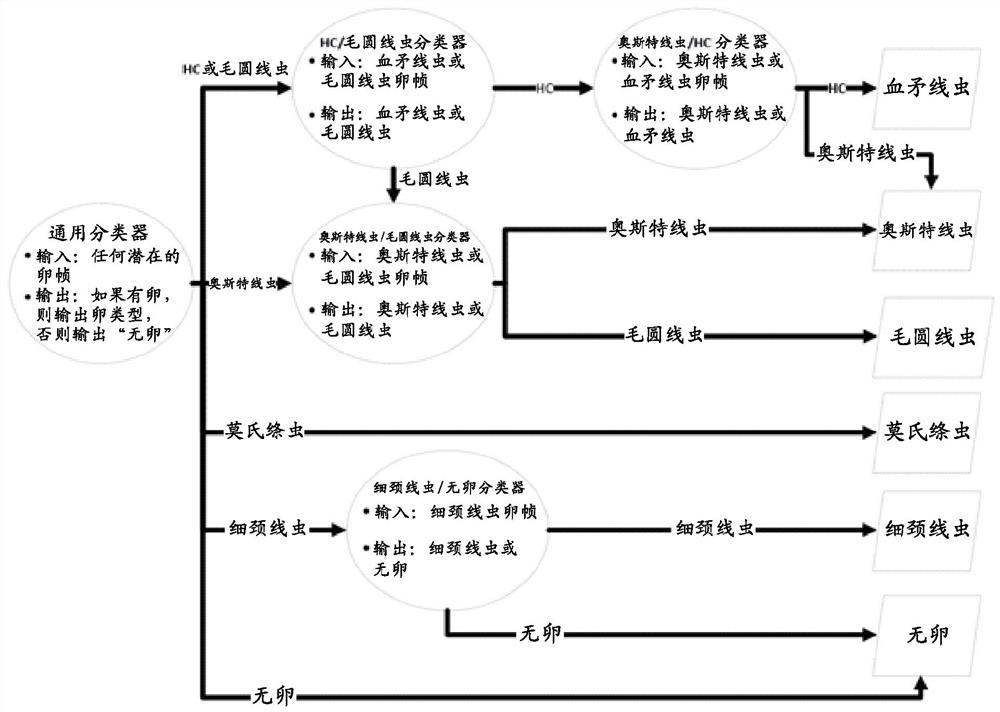
[0237] Example 1: Machine identification of parasite eggs
[0238] Parasite eggs are identified using a computer comprising a parasite egg identification algorithm according to the present invention. A total of 65 images were analyzed by computer for each of Haemonchus, Taenia mortensi, Leucocele, Osteria, and Trichostrongylus. The results of precision and recall are shown in Table 2 below:
[0239]
[0240] It is conceivable that the performance of the algorithm could be improved by training with a larger number of training images and / or a more diverse range of training images.
example 2
[0241] Example 2: Comparison of crop-fitting and template-based methods during recognition
[0242] Using the clip-fit method versus the Performance of the detection algorithm for the template approach. The results are shown in Table 3 below:
[0243]
[0244] It is to be noted that the crop fitting method is generally superior.
example 3
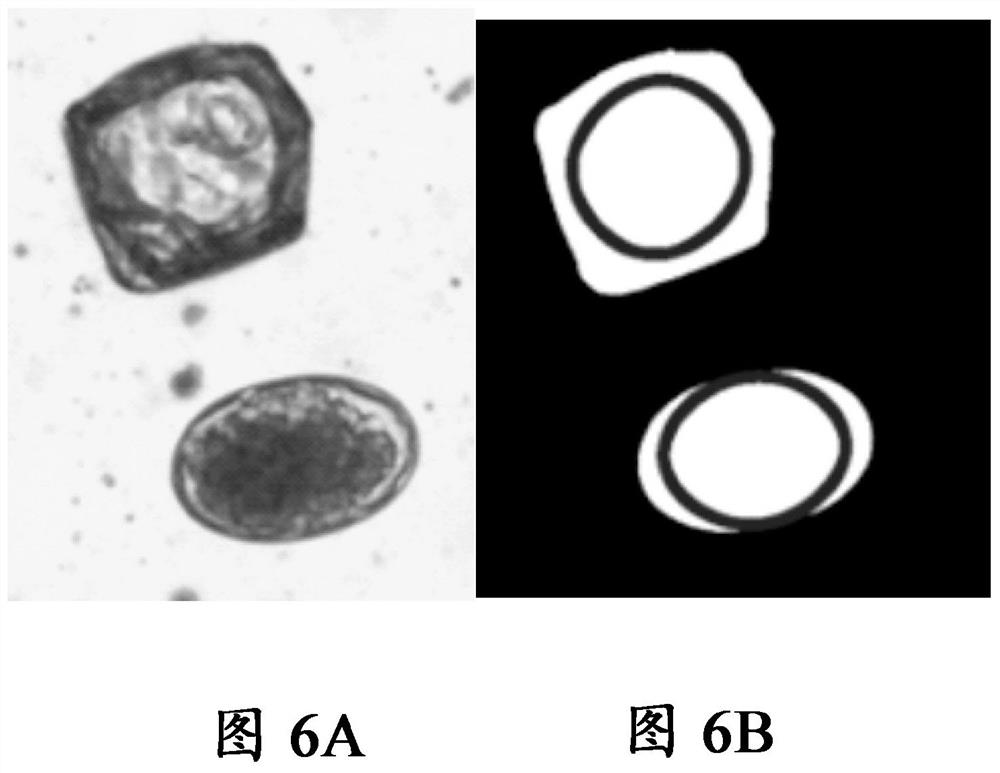
[0245] Example 3: Comparison of crop-fit and template-based methods in the identification of individual eggs and egg clusters
[0246] For each of Haemonchus (HC), Taenia mortensi (Mon), Nematode elegans (Nem), Oster and Trichostrongylus (Trich), with or without egg contact , evaluated the performance of detection algorithms using crop-fitting methods versus template-based methods. The results are shown in Table 4 below:
[0247]
[0248] It is generally noted that the crop-fit method outperforms the template method when a single egg is involved, whereas the opposite is observed when eggs are in contact.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com