Genetic disease high-throughput sequencing pathogenic mutation screening method based on core family
A screening method and a genetic disease technology, applied in the application and device fields of the above screening method, can solve the problems of being prone to errors and omissions, time-consuming and labor-intensive, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 2
[0070] Example 1 Genotype Display Mode in Next Generation Sequencing
[0071] Genotype refers to the general term for the entire gene combination of an individual organism, reflecting the sum of the genetic material obtained by the organism from its parents. The genotype specifically used in genetics generally only represents the composition of alleles at individual or a few loci. For example, if the reference sequence of a certain SNP site is A, and the two alleles of the gene where the site is located in a person are A and T respectively, then the genotype of this person's site is A / T . The genotype contains a variety of mutation types, not only single base substitution, but also insertion, deletion, insertion / deletion, inversion and many other mutation types.
[0072] High-throughput sequencing technology, also known as next-generation sequencing technology, can determine the sequence of the target fragment, extract the sites different from the reference sequence, and rec...
Embodiment 2 2
[0114] Core Family Analysis in Example 2 Next Generation Sequencing
[0115] The core family analysis model for high-throughput sequencing of genetic disease-causing mutations is called the Trios model in English. The pathogenic mutations hidden under the disease phenotype confirm the origin of the disease. If high-throughput sequencing is performed on the proband, after the analysis finds a possible pathogenic mutation, and then the parents are verified by next-generation sequencing to determine the source of the pathogenic mutation in the proband, then this method of analysis is generally accepted. It is called proband analysis, not the core family analysis method mentioned in this article. More and more Trios family detection models have been used in domestic and foreign research literature on various genetic diseases in recent years. In 2014, an article was published on JAMA (Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, et al. Clinical exome seq...
Embodiment 3
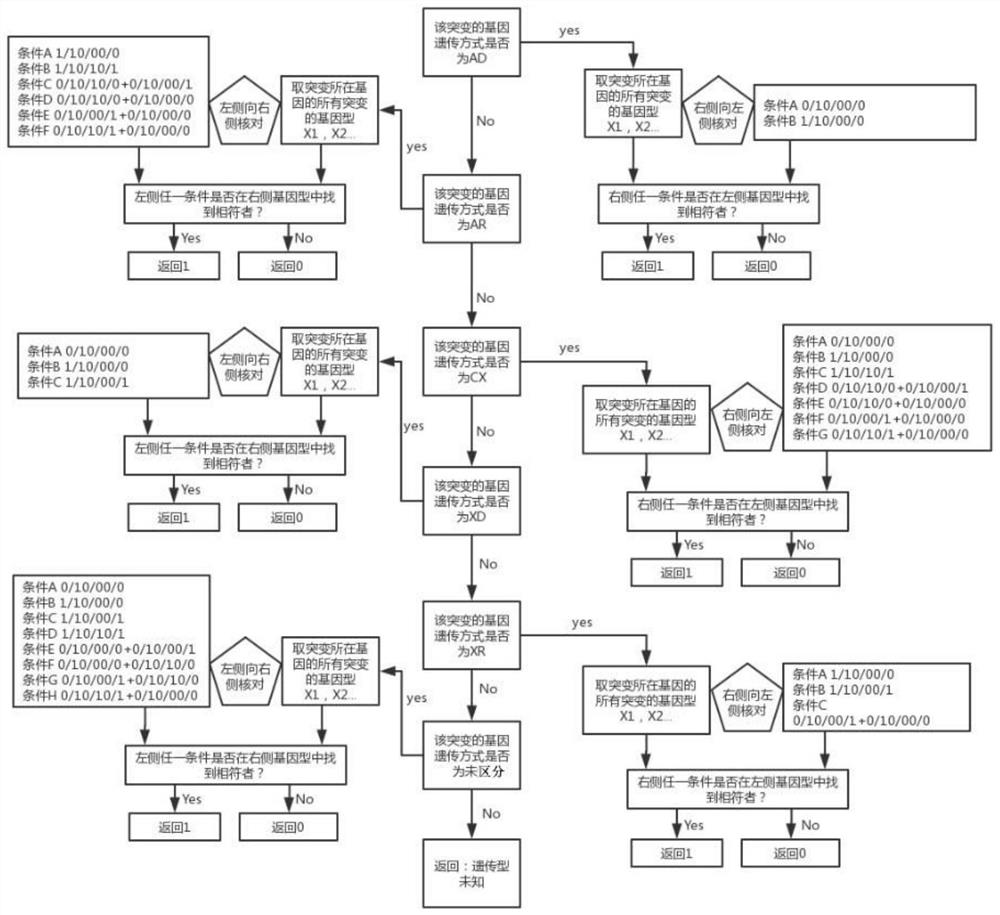
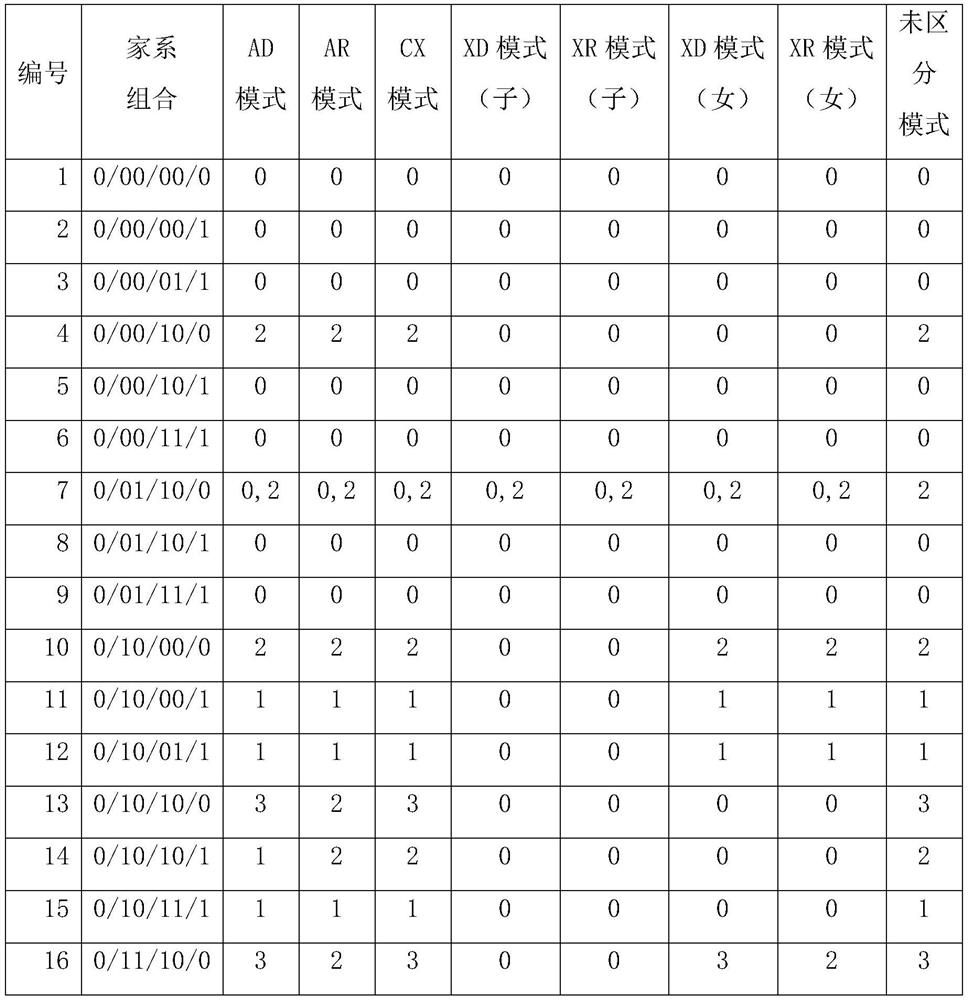
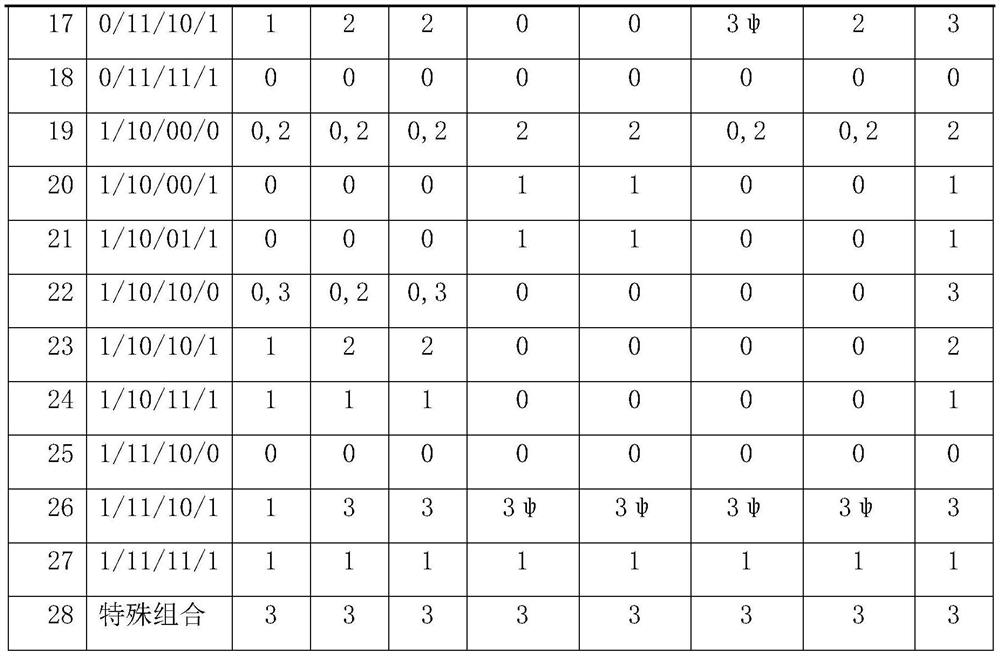
[0119] The family combination type of embodiment 3 mutation
[0120] As mentioned above, the genotype of a certain mutation site in the proband has three situations, 0 / 0, 0 / 1, 1 / 1, and the parents also have the same three situations. A certain mutant genotype is combined, in theory, there are 27 kinds. They are: 0 / 00 / 00 / 0; 0 / 00 / 00 / 1; 0 / 00 / 01 / 1; 0 / 00 / 10 / 0; 0 / 00 / 10 / 1; 0 / 00 / 11 / 1; 0 / 01 / 10 / 0; 0 / 01 / 10 / 1; 0 / 01 / 11 / 1; 0 / 10 / 00 / 0; 0 / 10 / 00 / 1; 0 / 10 / 01 / 1; 0 / 10 / 10 / 0; 0 / 10 / 10 / 1; 0 / 10 / 11 / 1; 0 / 11 / 10 / 0; 0 / 11 / 10 / 1; 0 / 11 / 11 / 1; 10 / 00 / 0; 1 / 10 / 00 / 1; 1 / 10 / 01 / 1; 1 / 10 / 10 / 0; 1 / 10 / 10 / 1; 1 / 10 / 11 / 1; 1 / 11 / 10 / 0; 1 / 11 / 10 / 1; 1 / 11 / 11 / 1; plus special combinations belonging to other situations, a total of 28 kinds.
[0121] In the present invention, data analysis is performed on 10 families that have undergone whole-exon high-throughput sequencing experiments. The data for each combination is as follows:
[0122]
[0123]
[0124]
[0125] It can be seen from the above table that during the next-g...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap