Compositions and methods for digital polymerase chain reaction
A polynucleotide, signal technology, applied in the field of sequence variants in nucleic acid samples before
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
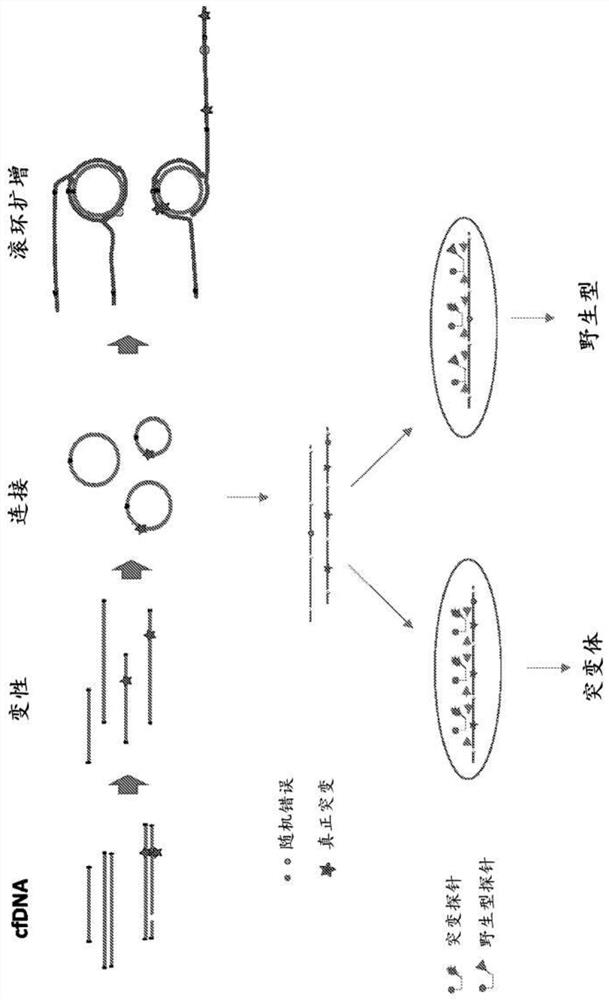
[0143] Example 1: ddPCR analysis of cancer variants using WGA amplified short fragmented cfDNA reference standard samples
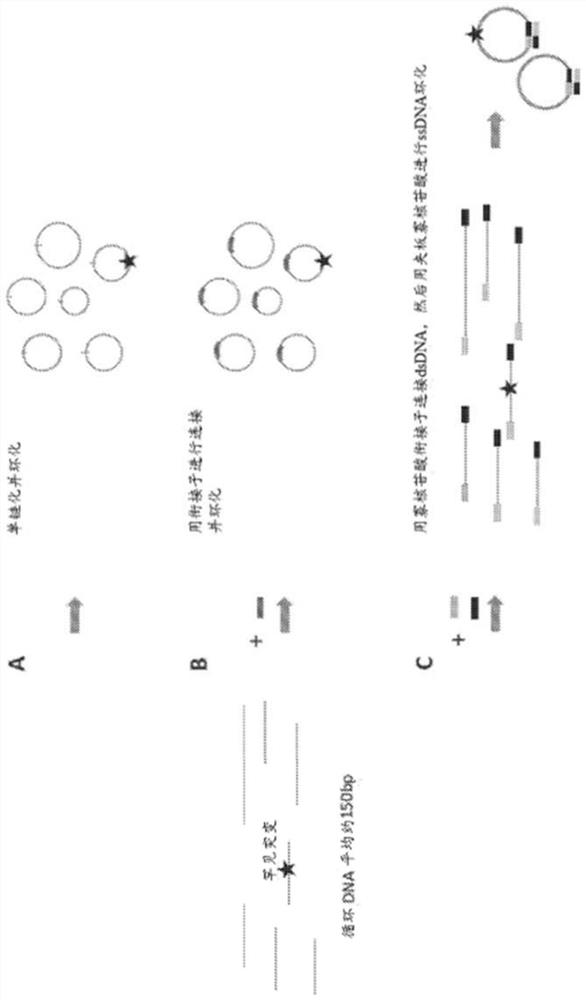
[0144] A cfDNA reference standard was prepared by mixing short fragmented DNA of approximately 150 bp in size from different cancer cell lines with NA12878 in different ratios. Four different cfDNA reference standards were used in this study: 5 ng 0.25% reference standard; 10 ng 0.25% reference standard; 20 ng 0.1% reference standard; and 20 ng 0% reference standard.
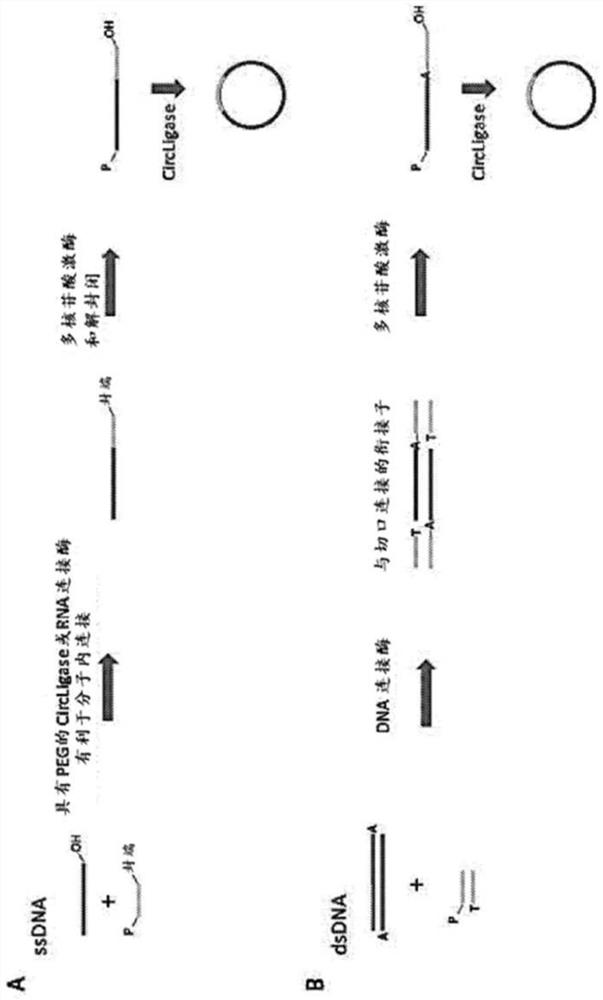
[0145] Each sample has 3 replicates. DNA samples were denatured at 96 °C for 30 s and cooled on ice for 2 min. Addition of ligation mix (2 μL 10x CircLigase buffer, 4 μL SM Betaine, 1 μL 50 mM MnCh, 1 μL Circligase II (Epicentre #CL9025K)) was established on a cold block and ligation was performed at 60°C for 3 hours. The ligated DNA mixture was incubated on a PCR machine at 80°C for 45 seconds, followed by exonuclease treatment. 1 μL of exonuclease mix (Exol 20U / μL:ExoIII 100U / μL=1:2) w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com