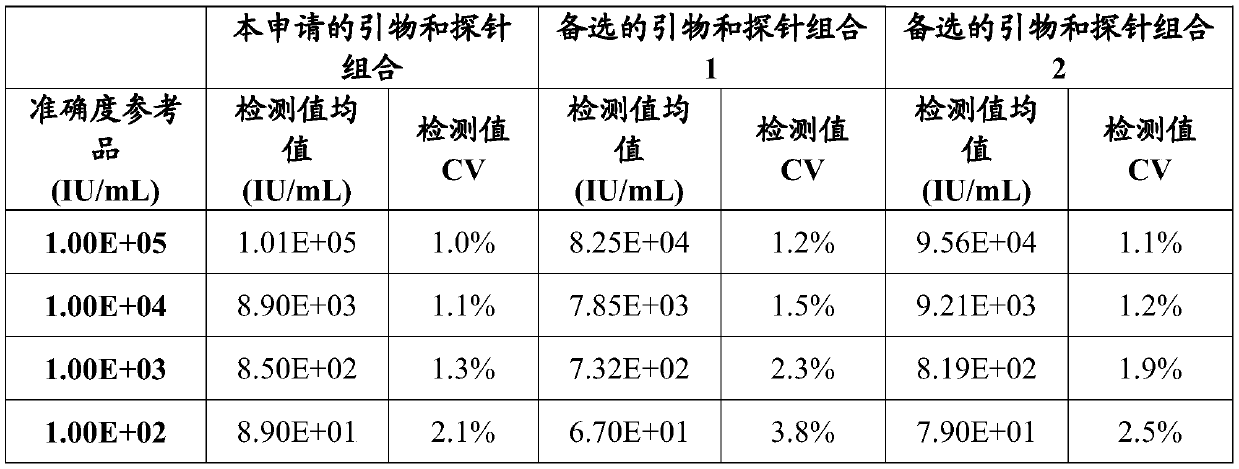
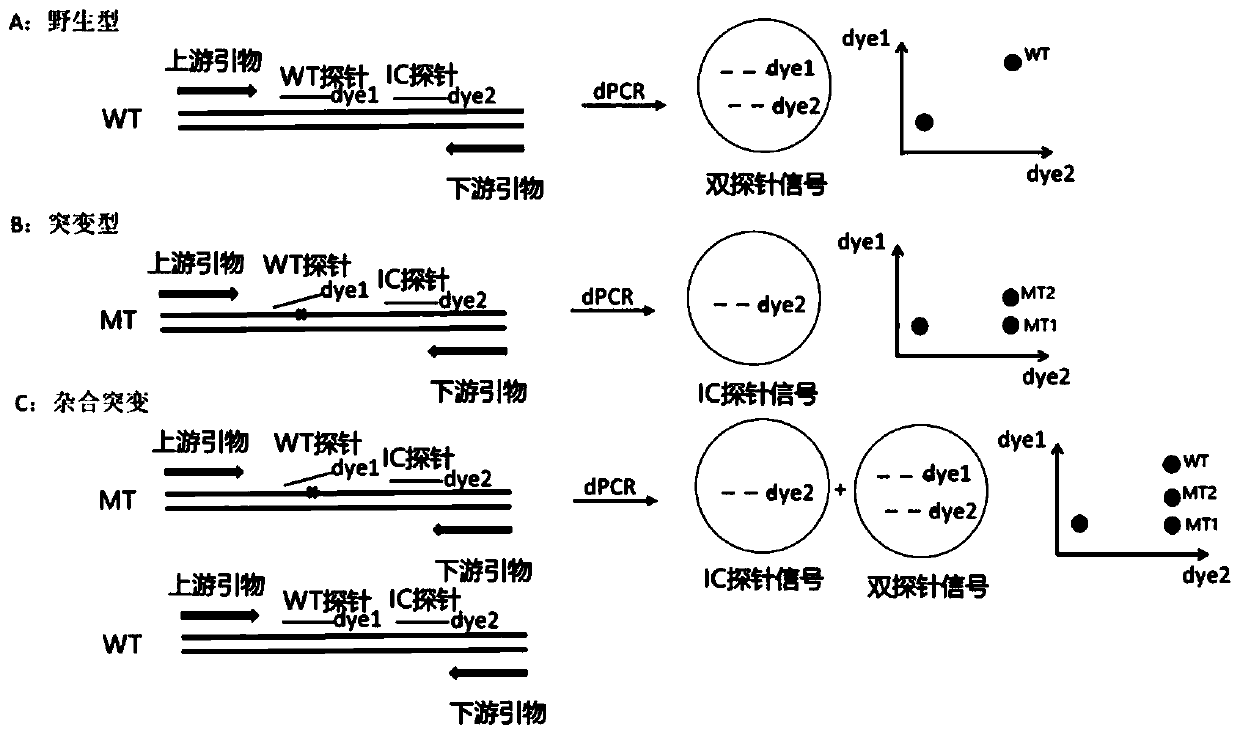
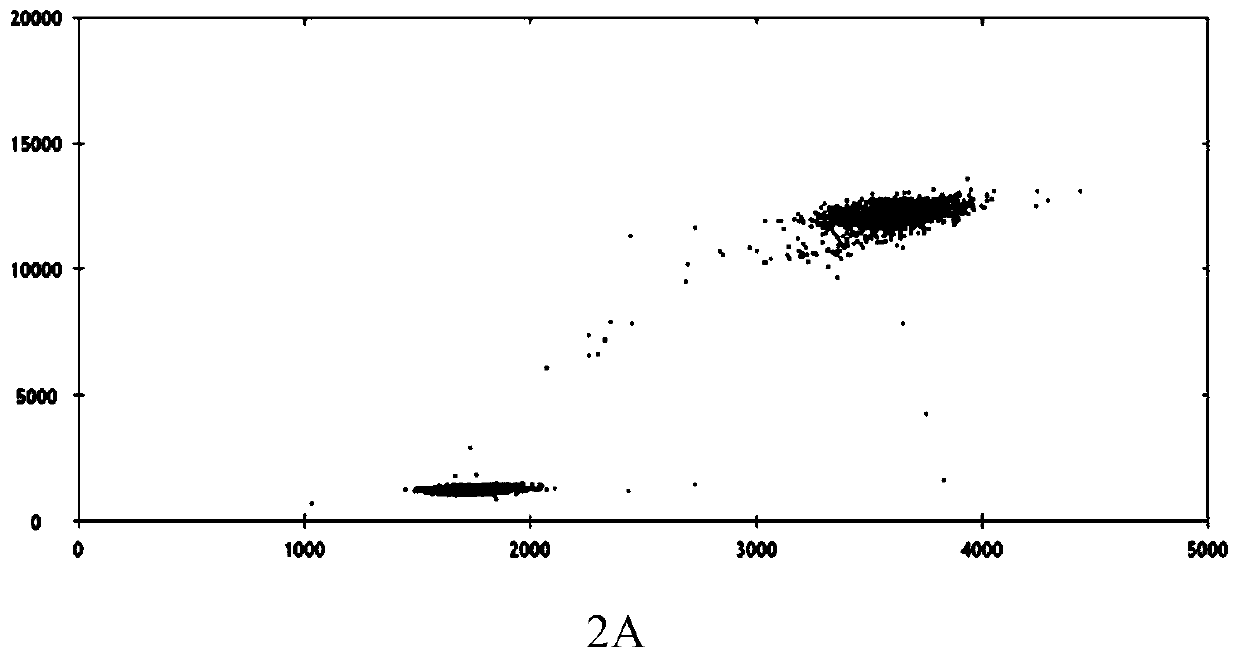
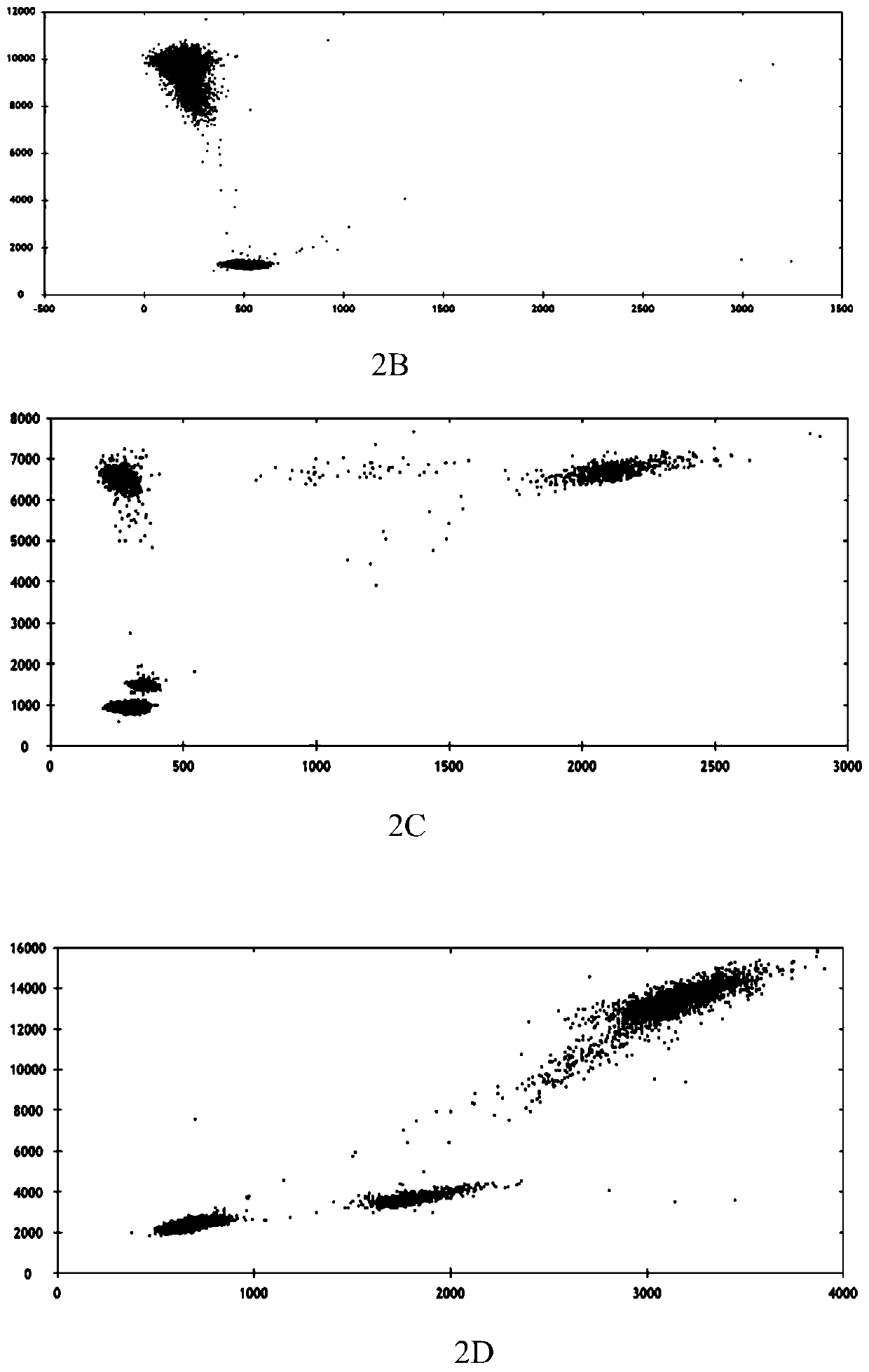
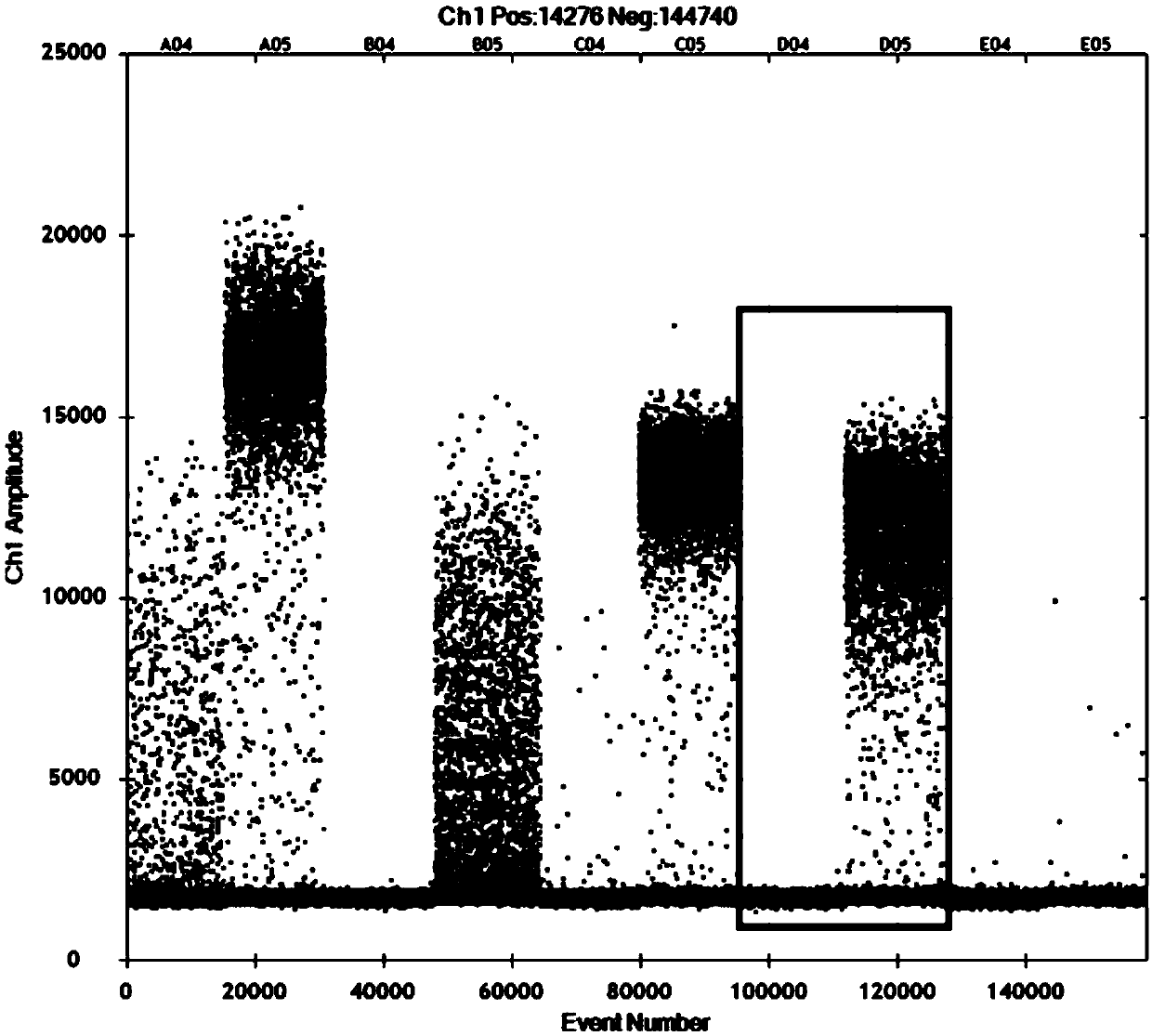
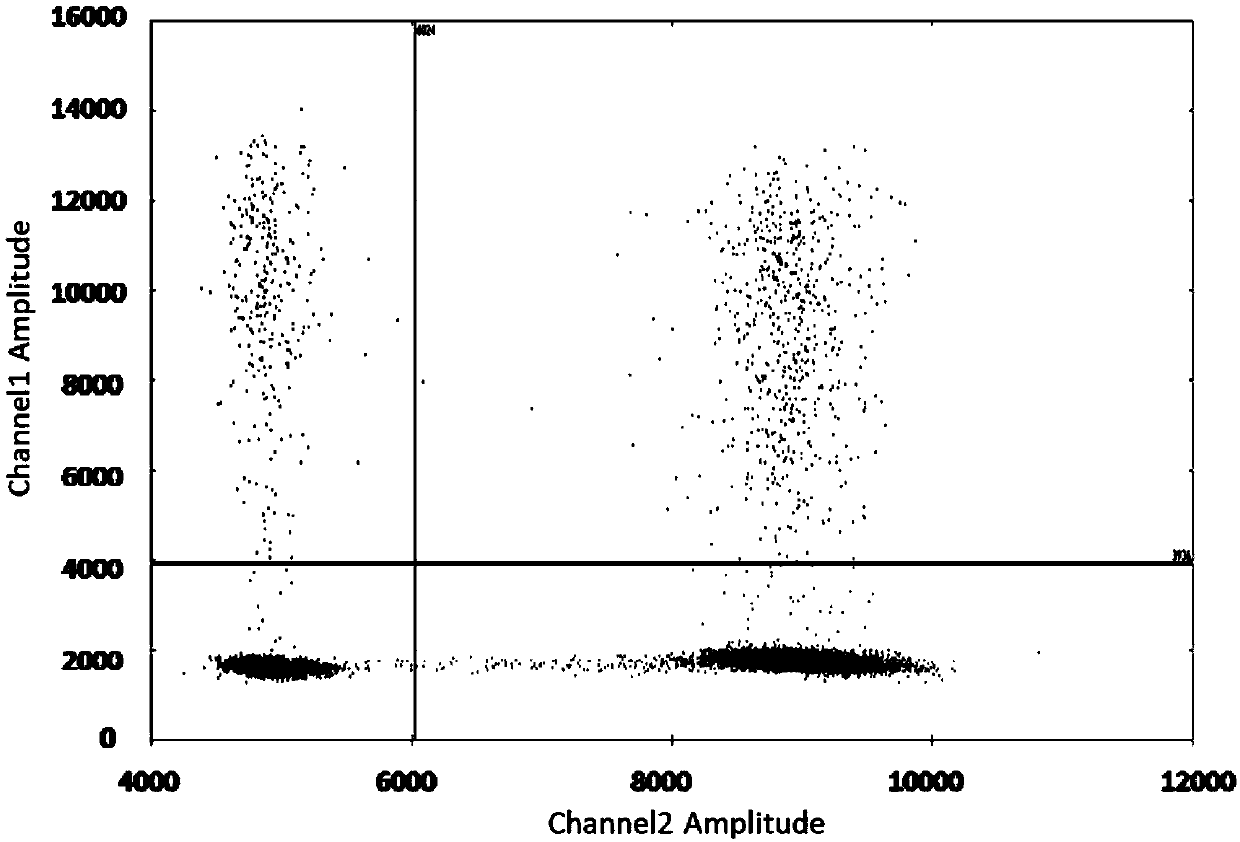
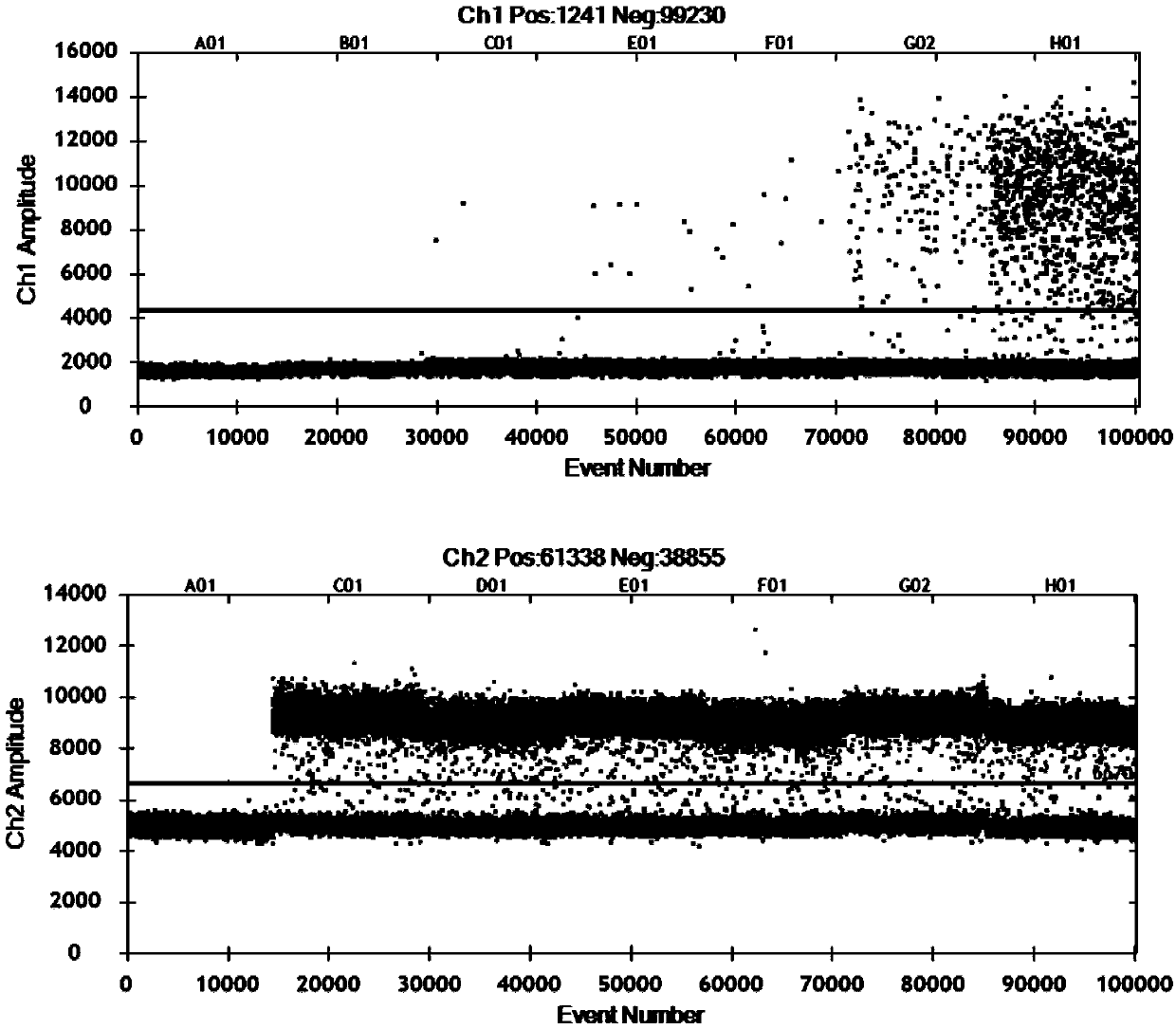
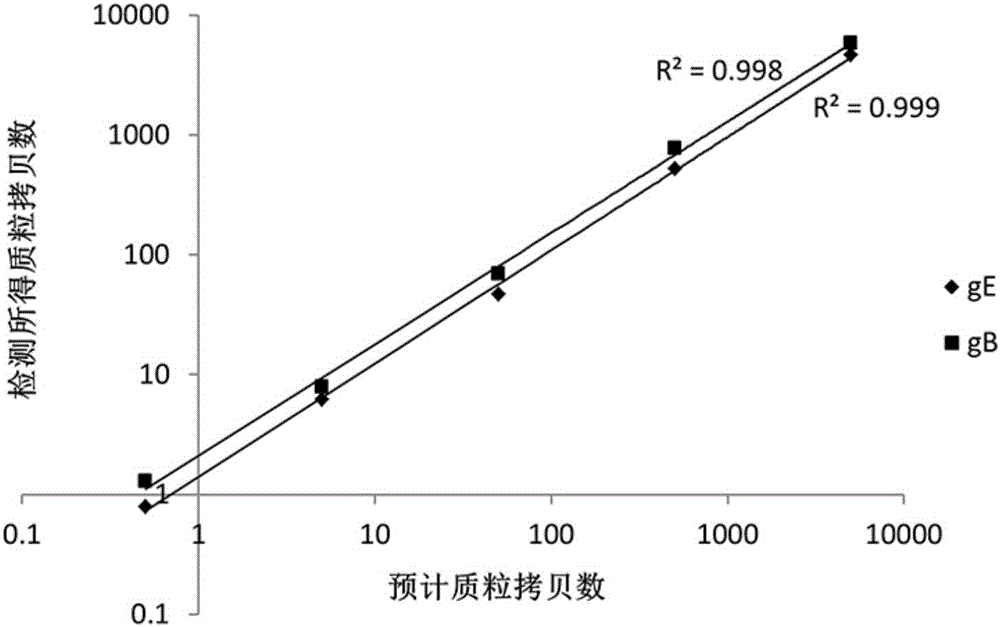
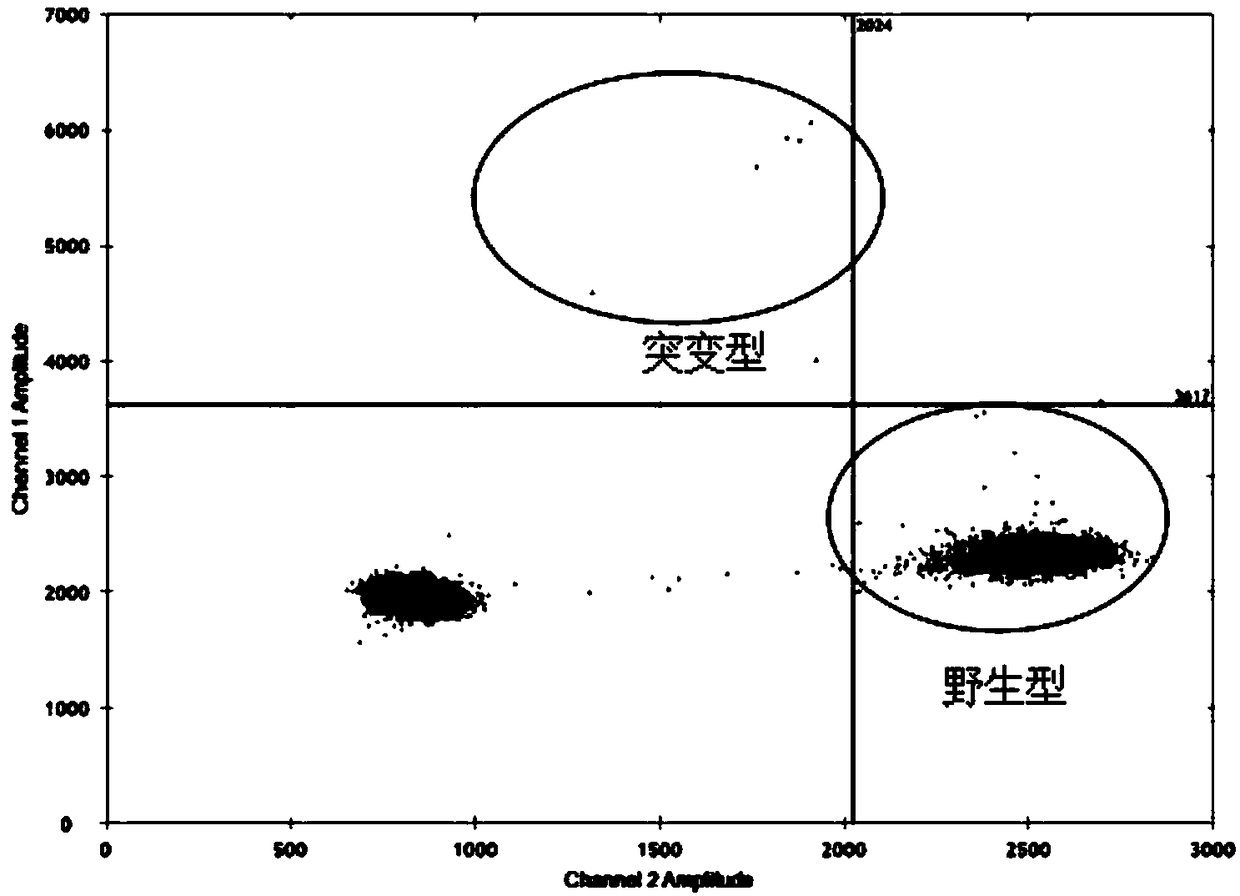
Patents
Literature
48 results about "Digital polymerase chain reaction" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
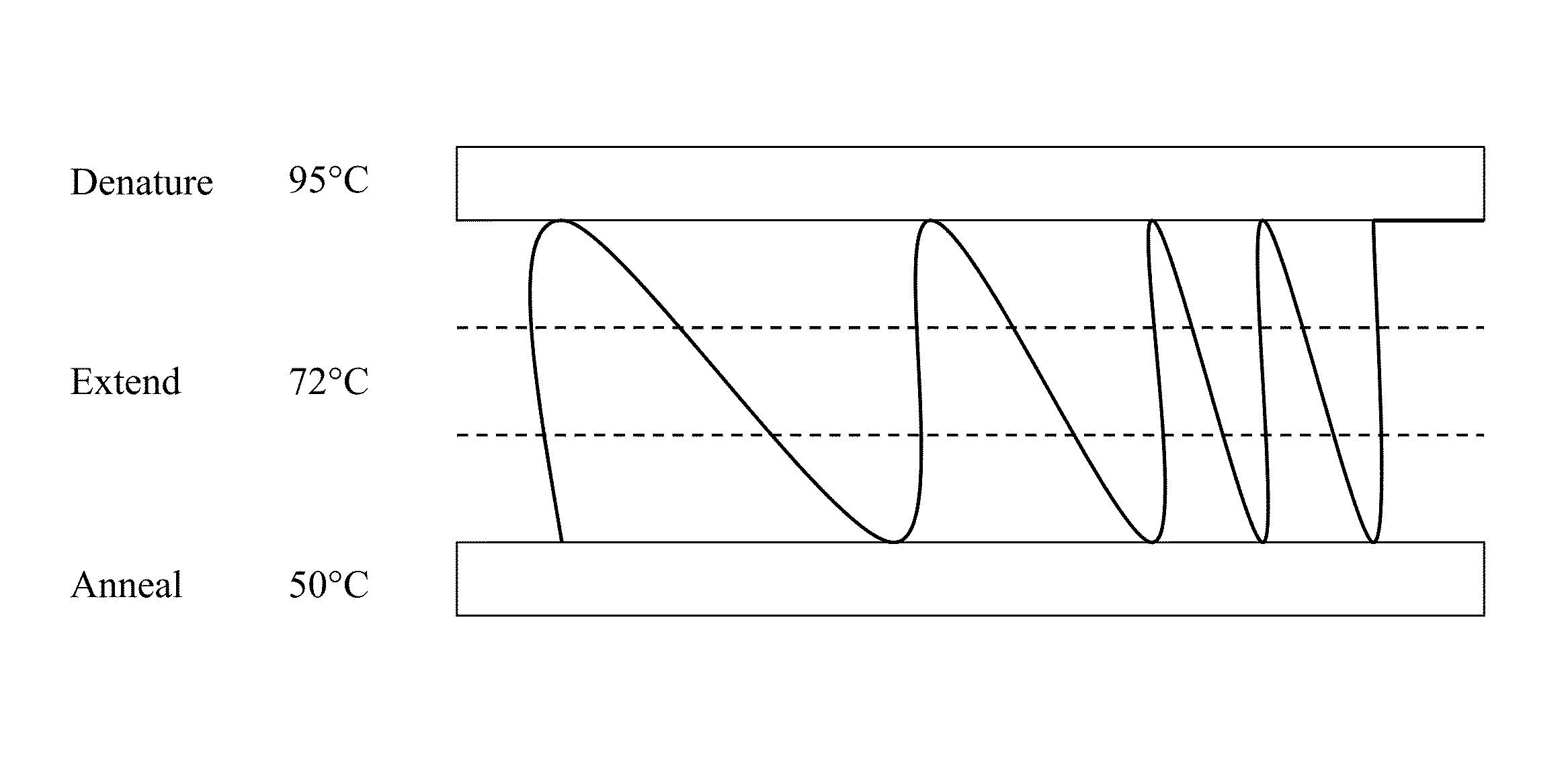
Digital polymerase chain reaction (digital PCR, DigitalPCR, dPCR, or dePCR) is a biotechnological refinement of conventional polymerase chain reaction methods that can be used to directly quantify and clonally amplify nucleic acids strands including DNA, cDNA or RNA. The key difference between dPCR and traditional PCR lies in the method of measuring nucleic acids amounts, with the former being a more precise method than PCR, though also more prone to error in the hands of inexperienced users. A "digital" measurement quantitatively and discretely measures a certain variable, whereas an “analog” measurement extrapolates certain measurements based on measured patterns. PCR carries out one reaction per single sample. dPCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. This separation allows a more reliable collection and sensitive measurement of nucleic acid amounts. The method has been demonstrated as useful for studying variations in gene sequences — such as copy number variants and point mutations — and it is routinely used for clonal amplification of samples for next-generation sequencing.
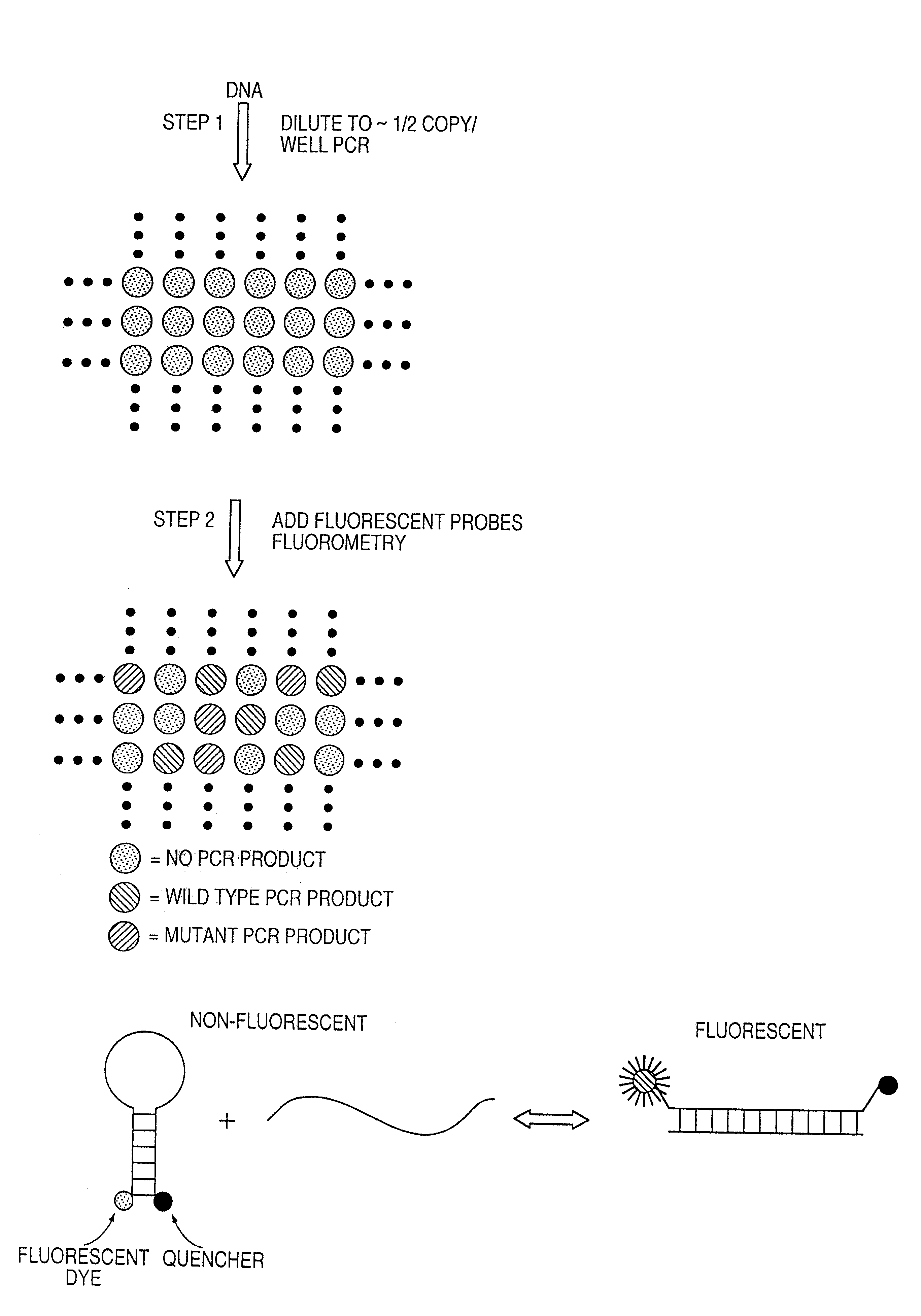
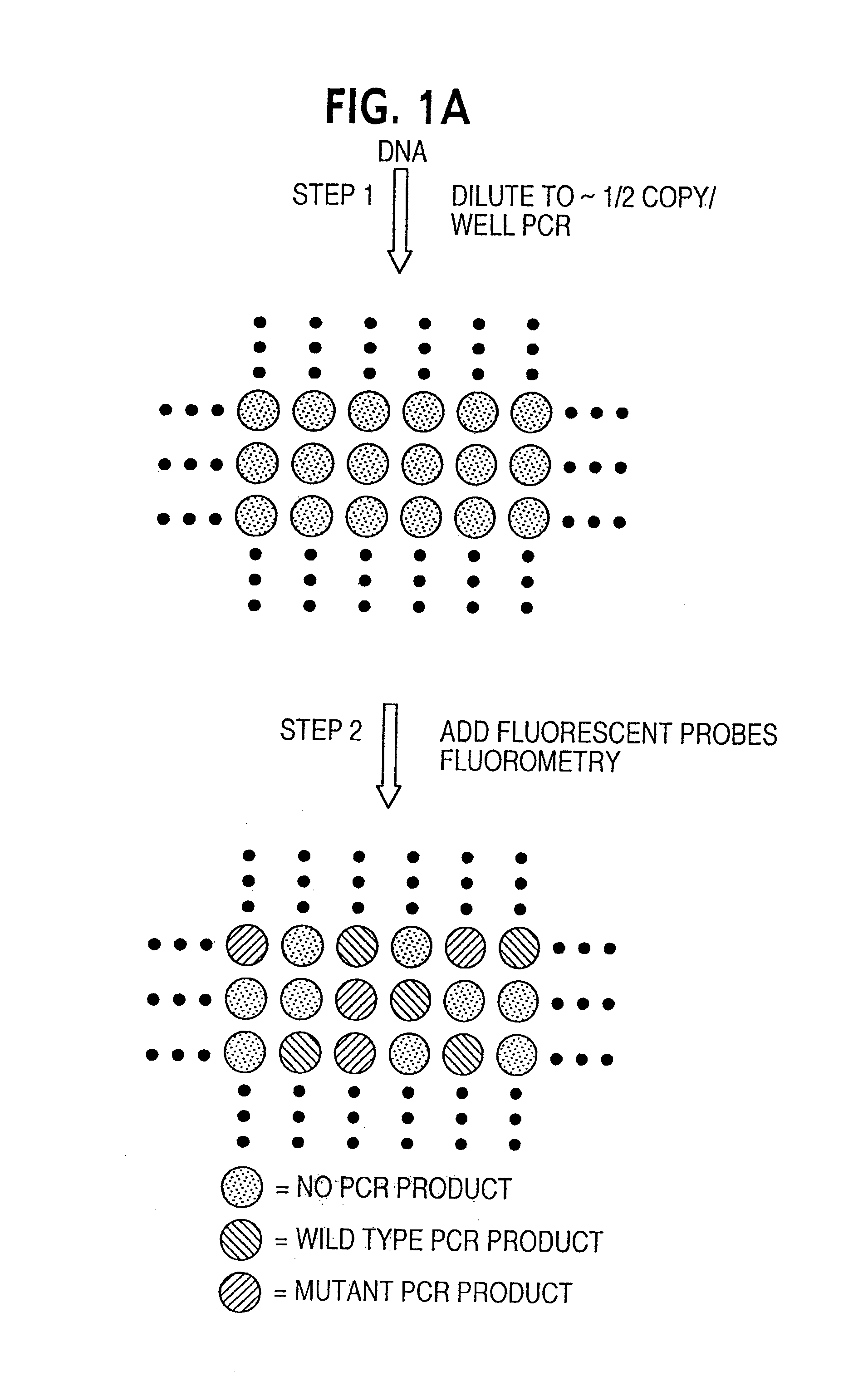
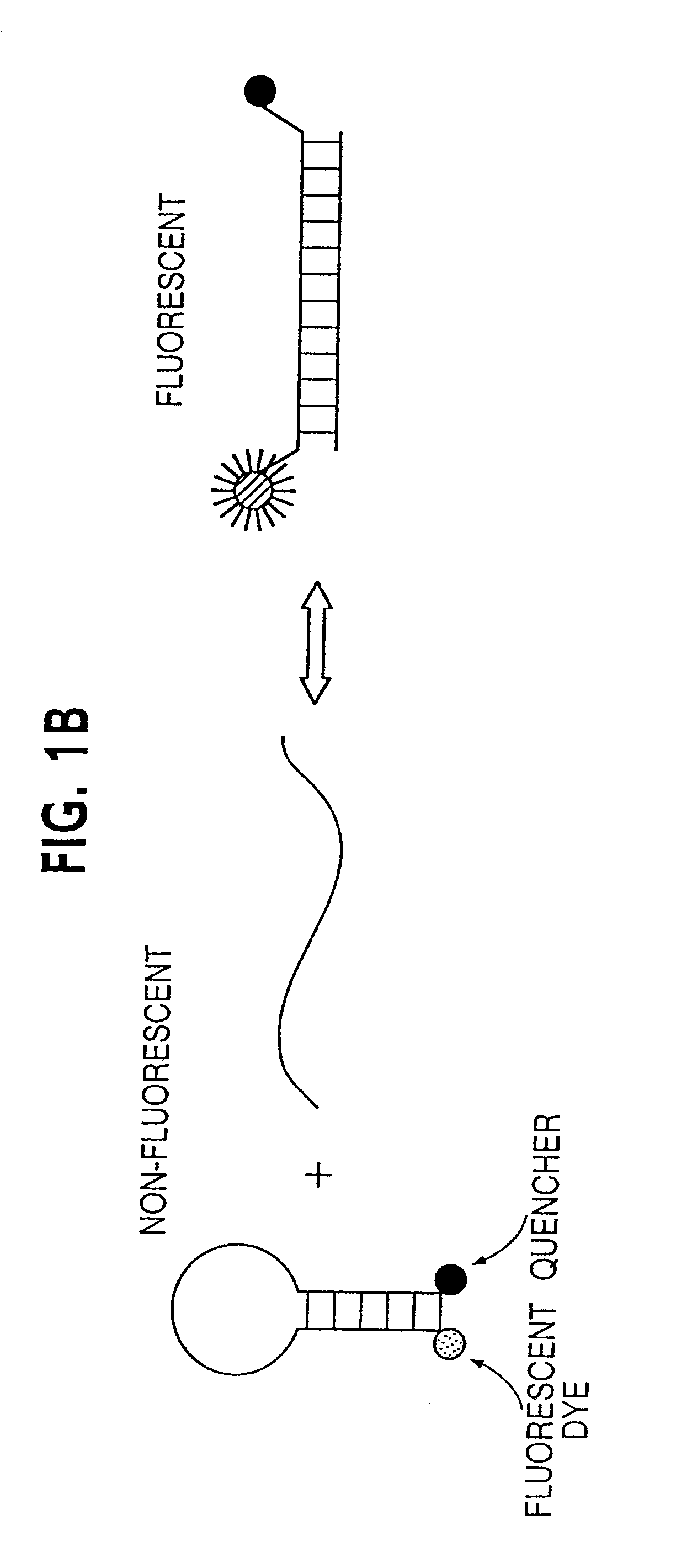
Digital amplification
The identification of pre-defined mutations expected to be present in a minor fraction of a cell population is important for a variety of basic research and clinical applications. The exponential, analog nature of the polymerase chain reaction is transformed into a linear, digital signal suitable for this purpose. Single molecules can be isolated by dilution and individually amplified; each product is then separately analyzed for the presence of pre-defined mutations. The process provides a reliable and quantitative measure of the proportion of variant sequences within a DNA sample.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
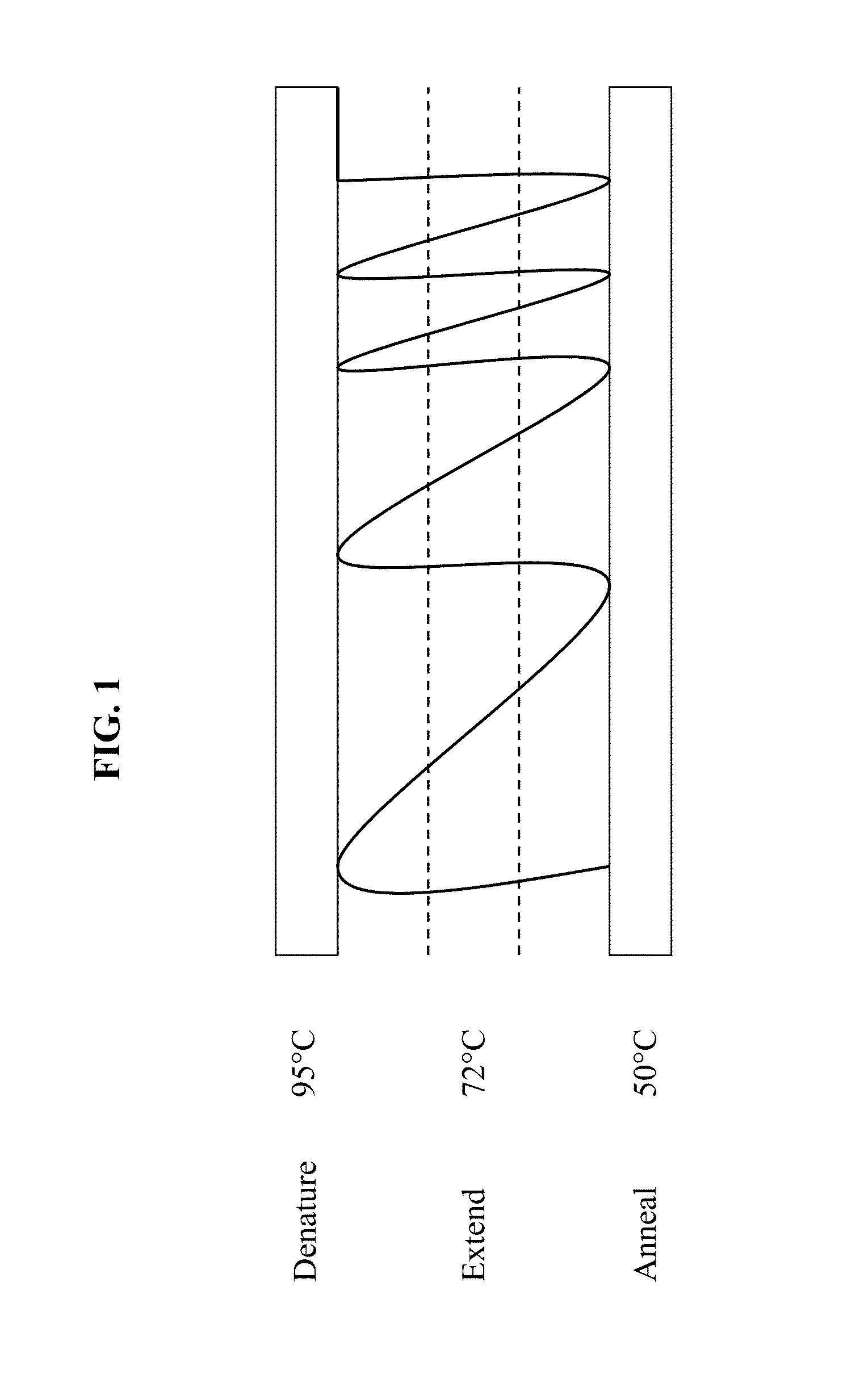
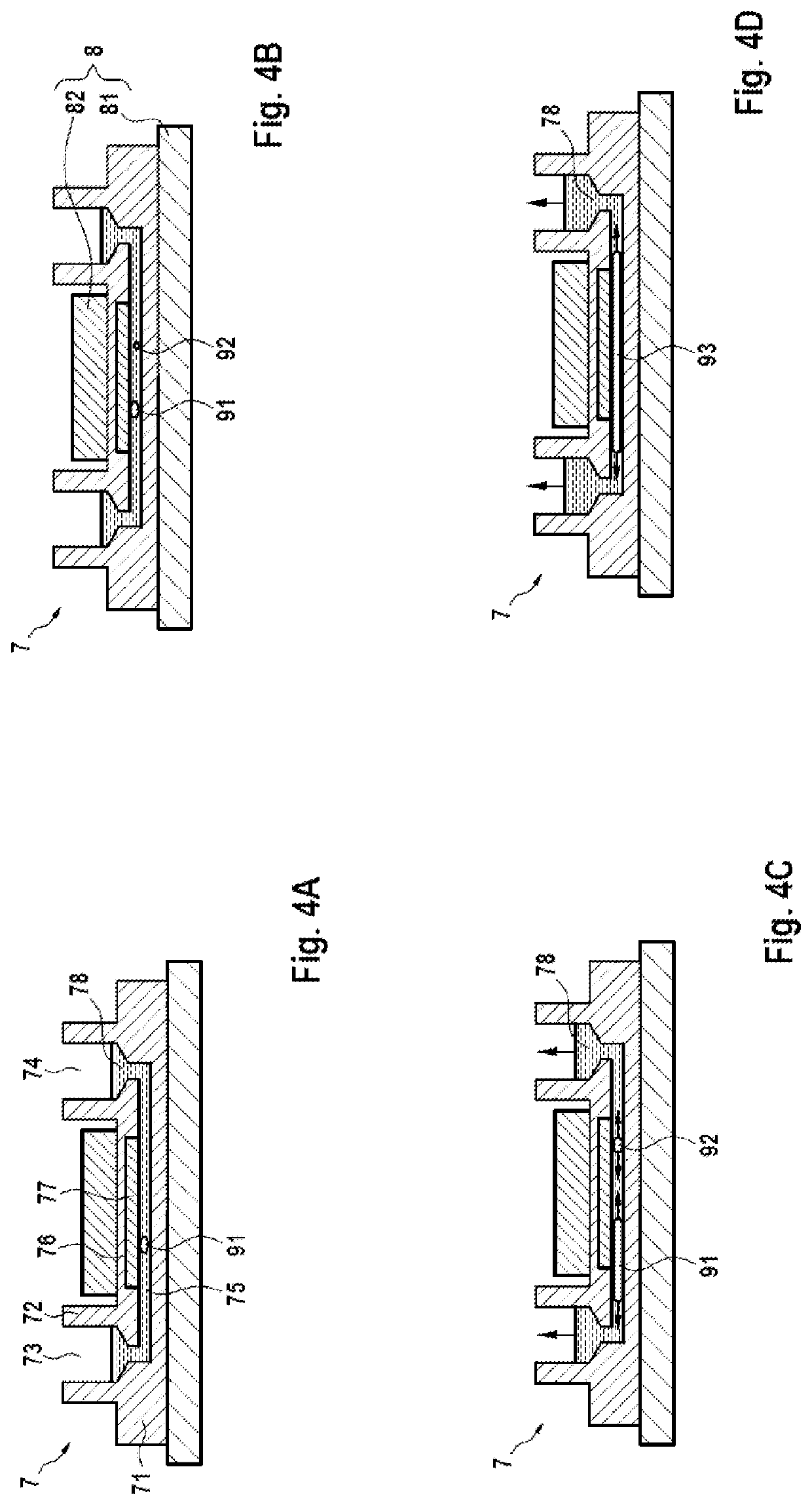
Channels with cross-sectional thermal gradients
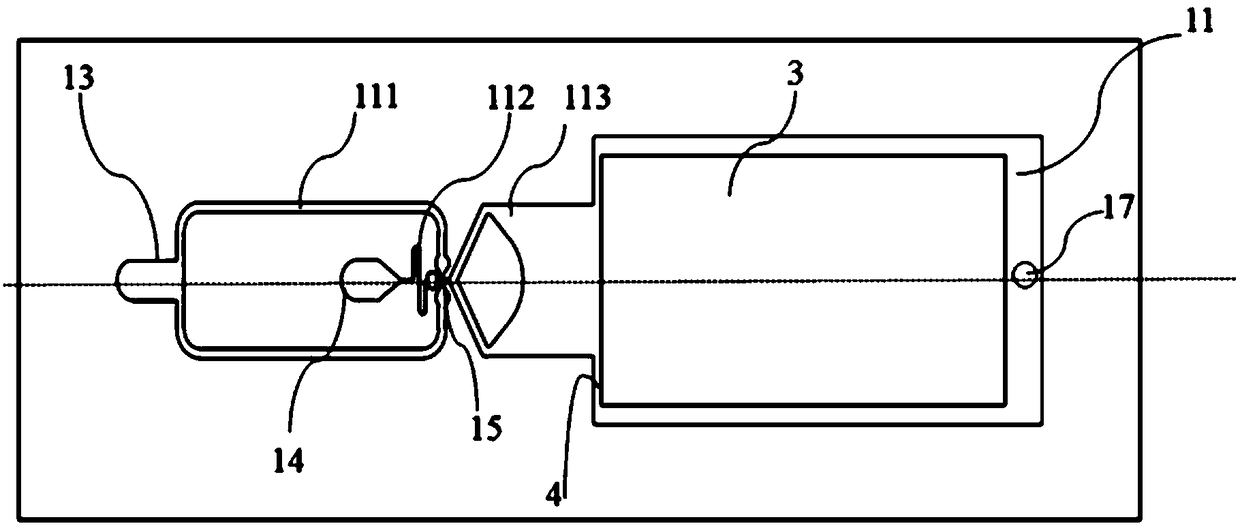
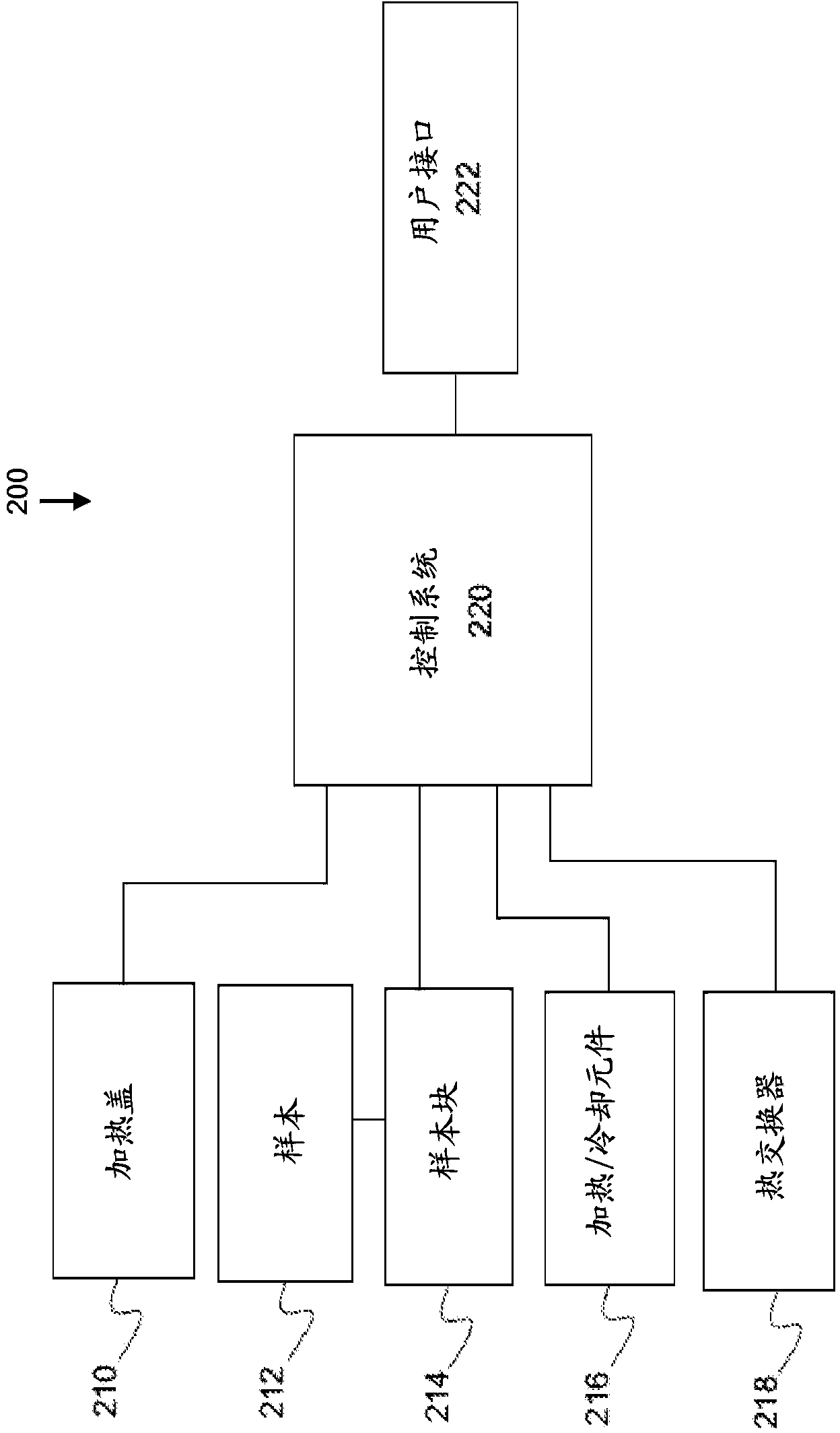
ActiveUS9222115B2Bioreactor/fermenter combinationsBiological substance pretreatmentsNucleic Acid DenaturationPolymerization
Owner:ABBOTT MOLECULAR INC
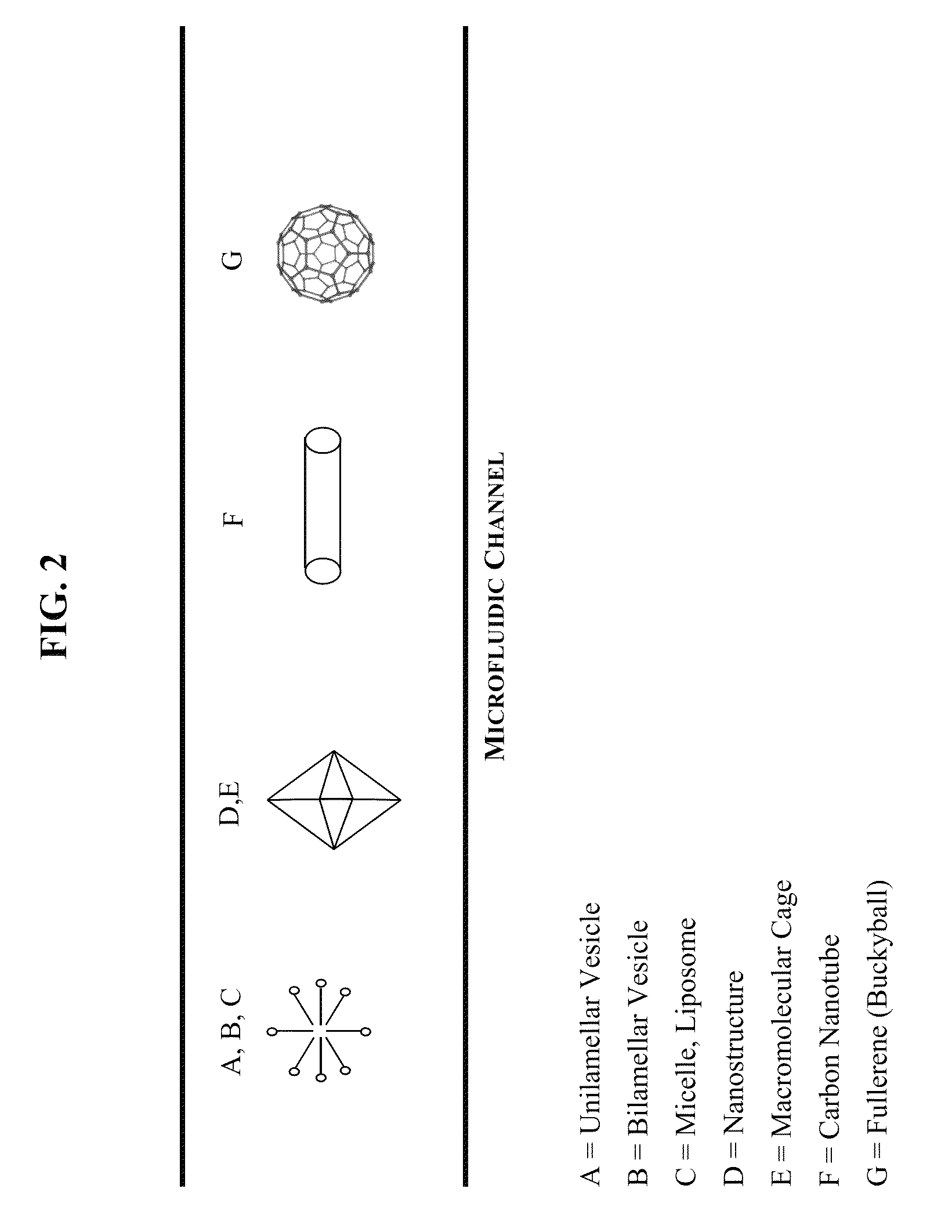
Nucleic acid detection micro-fluidic chip and preparation method thereof
InactiveCN109112063AAvoid fusesAvoid demulsificationBioreactor/fermenter combinationsBiological substance pretreatmentsNucleic acid detectionHigh volume manufacturing
The invention relates to a nucleic acid detection micro-fluidic chip, which comprises a substrate and a cover plate, wherein the cover plate is fixedly arranged on the substrate; the cover plate is provided with an oil phase sample-injection hole and a sample sample-injection hole; the surface, abutted with the substrate, of the cover plate is a structural surface; a first micro-flow channel connected with the oil phase sample-injection hole, a second micro-flow channel connected with the sample sample-injection hole and a liquid drop generation interface located in the joint of the first andsecond micro-flow channels, which are mutually communicated, are etched in the structural surface; the structural surface, in a liquid drop collection area, of the cover plate is fixedly provided witha thin plate; a gap is formed between the periphery of the thin plate and the boundary of the liquid drop collection area, so as to form a gas path; and the cover plate is also provided with a gas vent hole communicated with the gas path. The problems of the fusion and the demulsification of liquid drops are avoided; the homogeneity and the stability of the liquid drop are improved; meanwhile, the signal reading speed of the liquid drop is quickened during dPCR (digital Polymerase Chain Reaction) detection; and the detection period is shortened. The invention also relates to a preparation method of the nucleic acid detection micro-fluidic chip. The preparation method is simple and convenient; the cost is low; and the nucleic acid detection micro-fluidic chip not only can be produced in alarge-batch manner, but also can be produced in a laboratory in a small-batch manner.
Owner:北京旌微医学工程研究院有限公司
DPCR (Digital Polymerase Chain Reaction)-based novel full-automatic fluorescent signal acquisition and analysis method
ActiveCN107723344AAvoid interferenceHigh precisionMicrobiological testing/measurementFluorescence/phosphorescenceObservational errorImaging processing
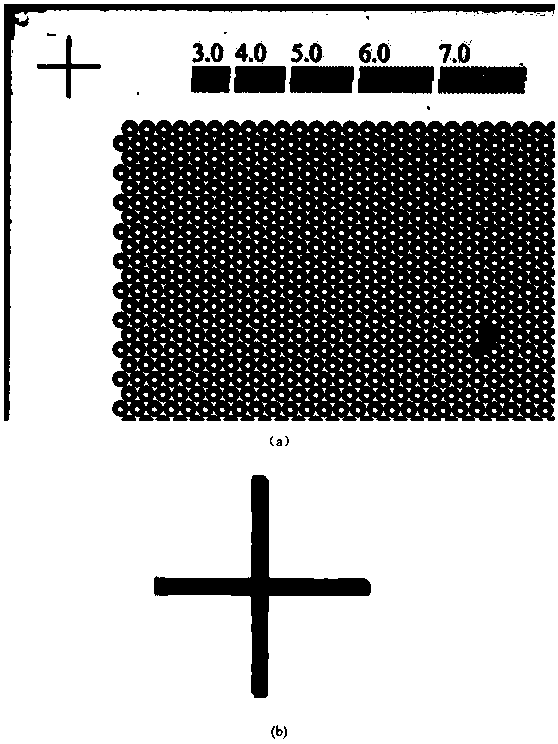
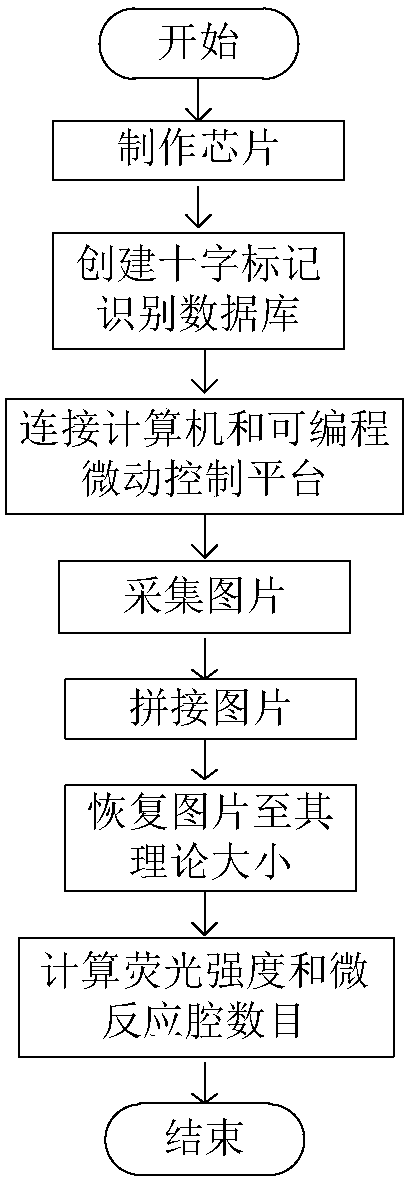
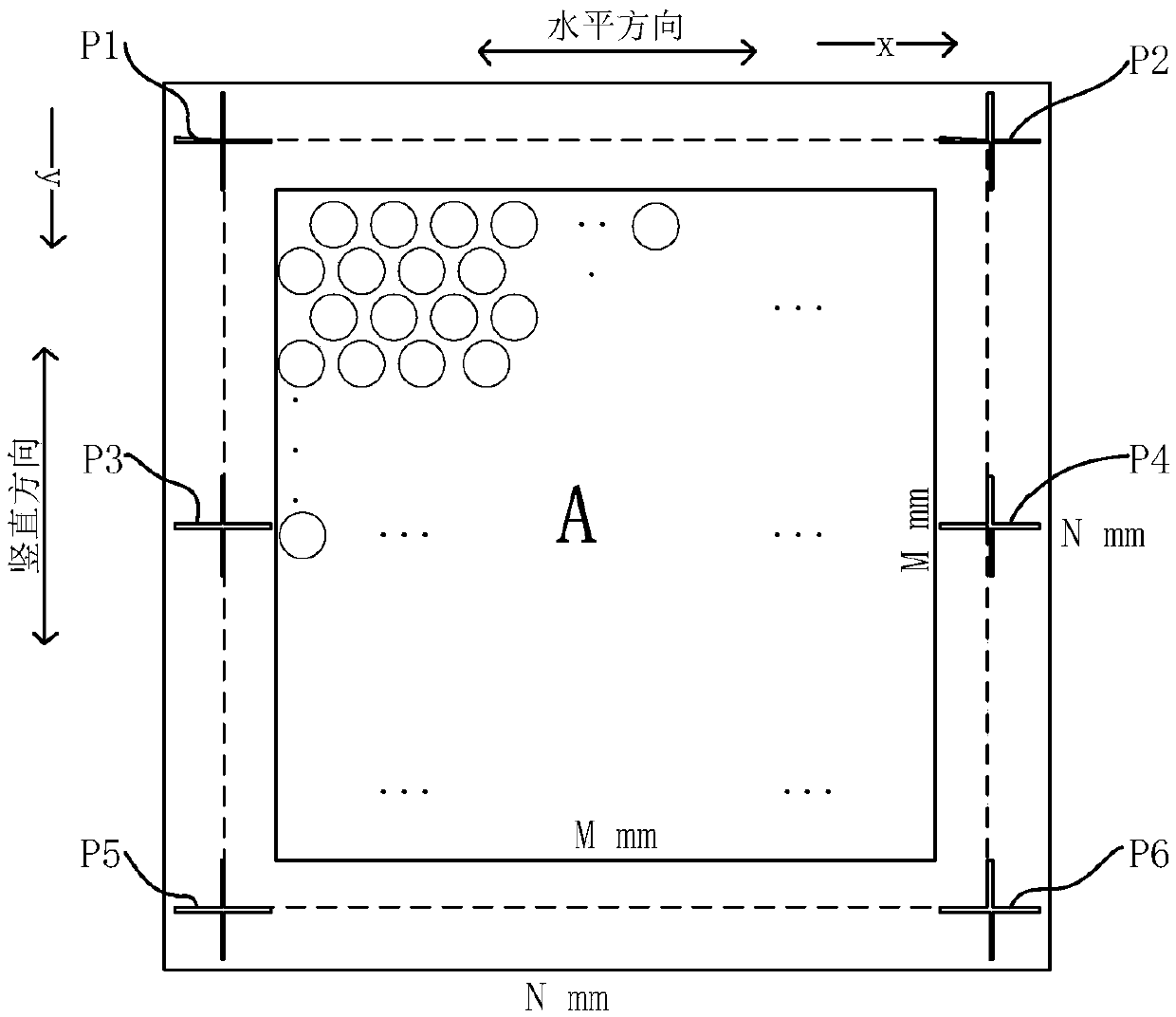
The invention belongs to the field of image processing and in particular discloses a dPCR (Digital Polymerase Chain Reaction)-based novel full-automatic fluorescent signal acquisition and analysis method. According to the method, a fluorescent signal is extracted and analyzed by utilizing an idea of image region segmentation-image stitching-signal acquisition and analysis. Due to the method, the fluorescent signal acquisition and analysis method is applied to result detection at the later stage of dPCR in an engineered manner firstly, and due to a reasonable design, full automation of the whole system is realized, measurement errors caused by artificial interference such as manual marking, progressive scanning and the like are avoided, the accuracy is high, the operation and control are convenient, and the detection precision and the detection efficiency of the system are effectively improved. Due to the stitching step, the acquired scattered images can be perfectly stitched to be of the size of the original image during designing, the original image is highly reduced, convenience is brought to fluorescent signal analysis and detection at the later stage, and the detection accuracyis improved.
Owner:NORTHWESTERN POLYTECHNICAL UNIV
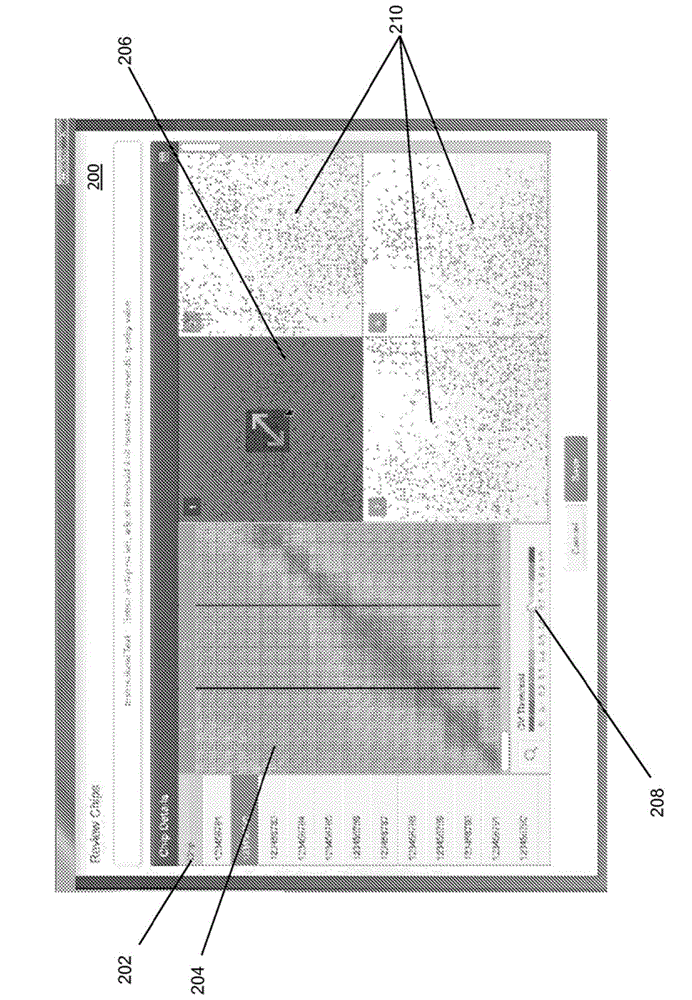
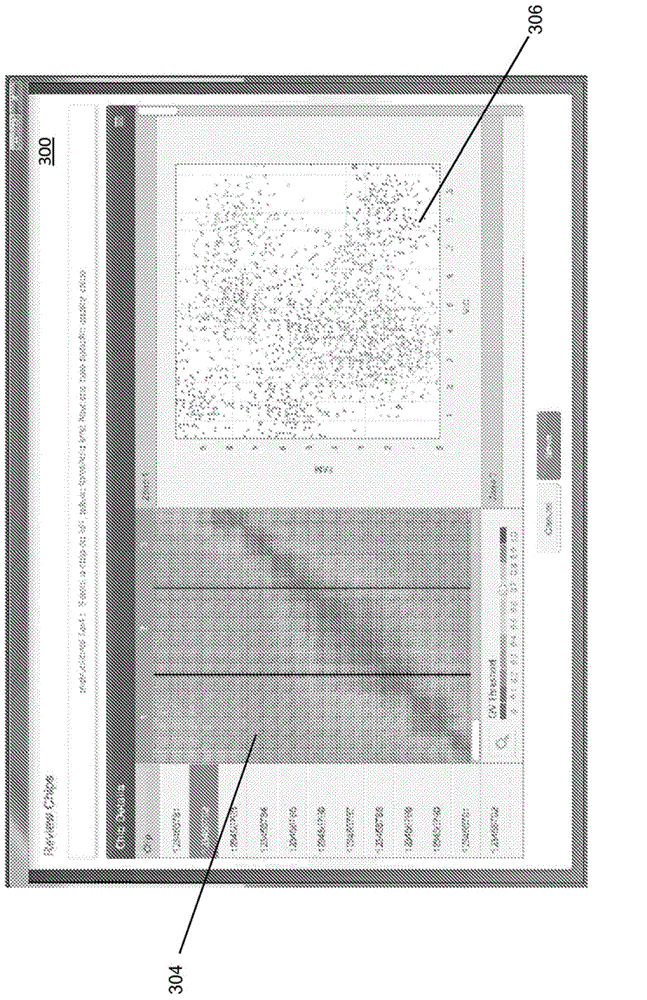
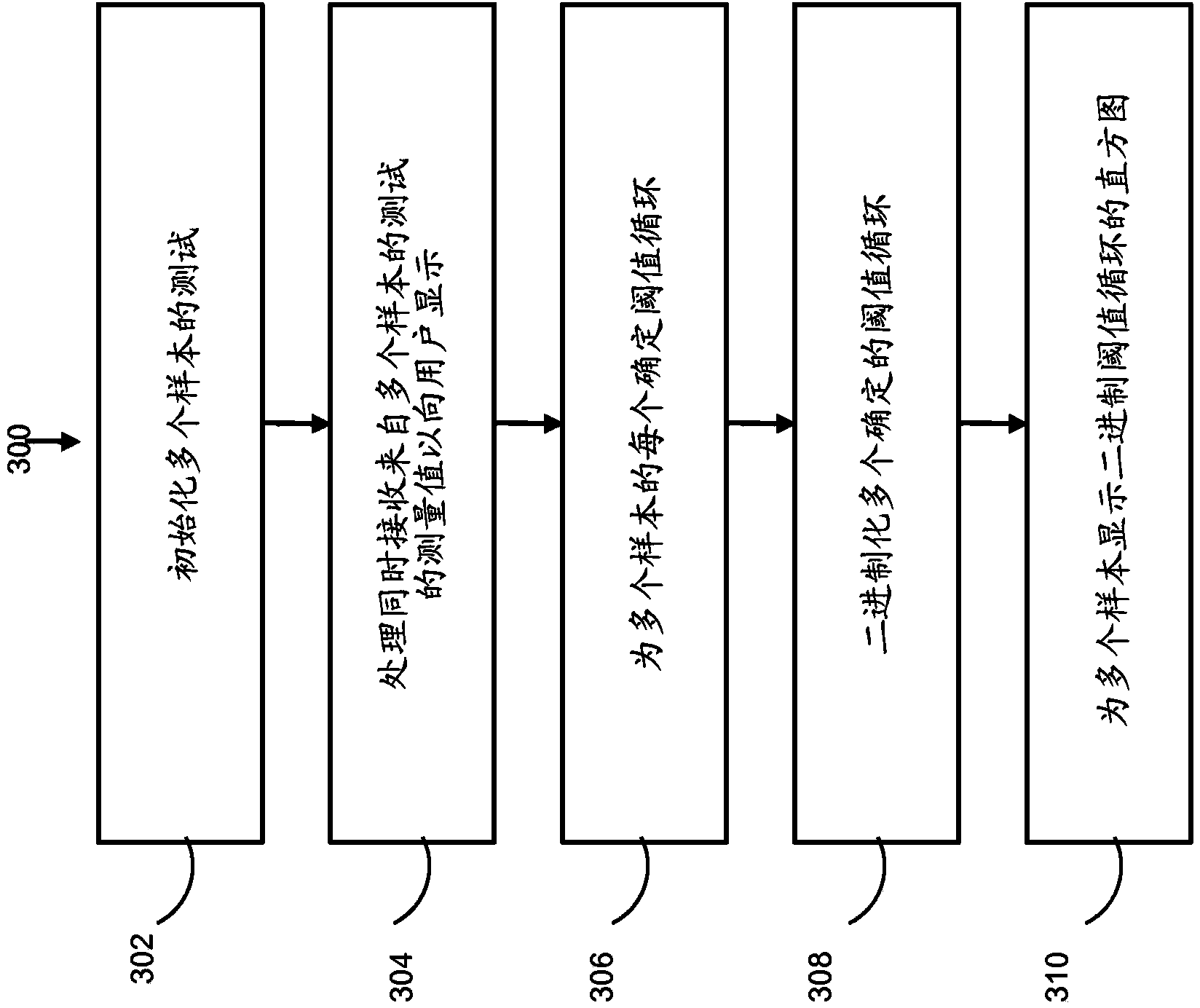
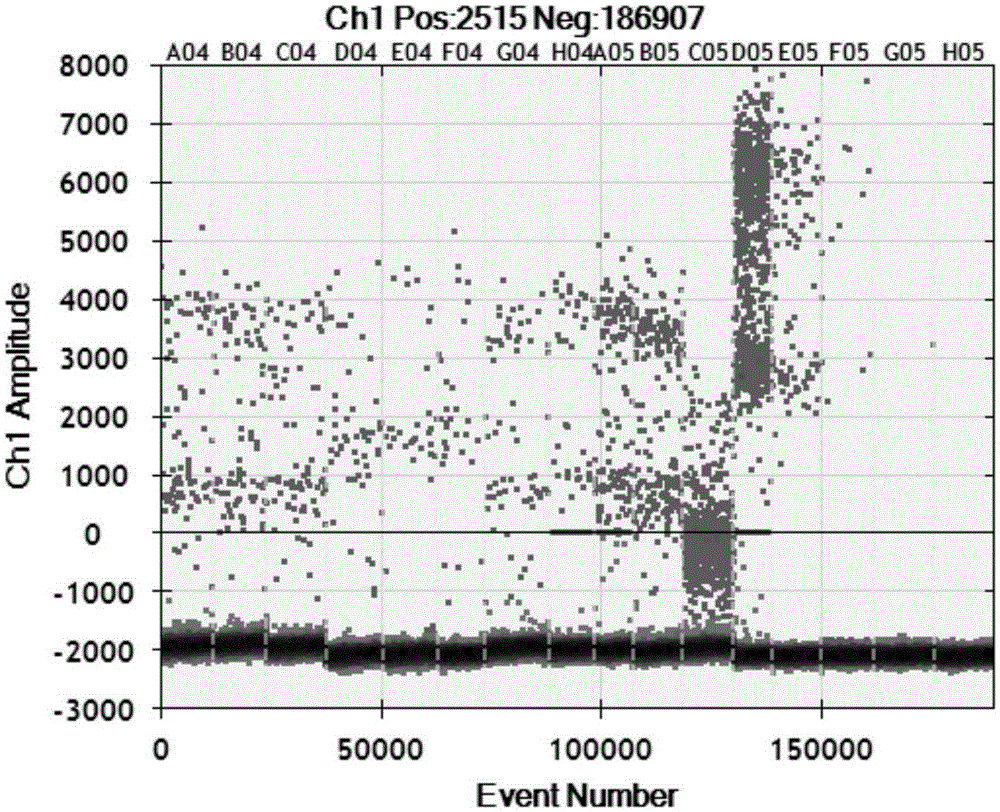
Visualization tools for digital polymerase chain reaction (PCR) data
A method for generating a data visualization is provided. The method includes displaying a representation of a portion of detected data from a substrate to a user. The method further includes generating a data quality value for the portion of detected data and displaying, along with the representation of the portion of detected data, an indication of data quality value for the portion of detected data. The method further includes selecting, by the user, a quality value threshold, and displaying an adjusted indication of data quality value for the portion of detected data meeting the quality value threshold.
Owner:LIFE TECH CORP
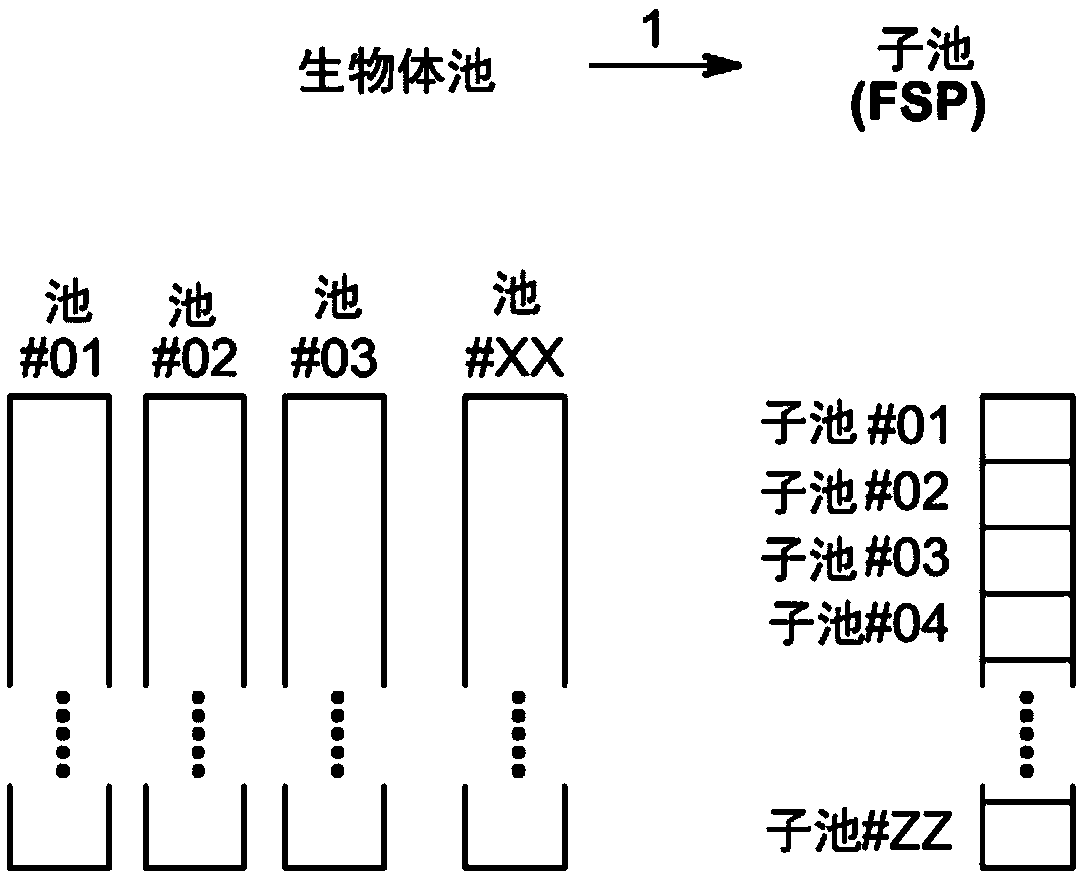
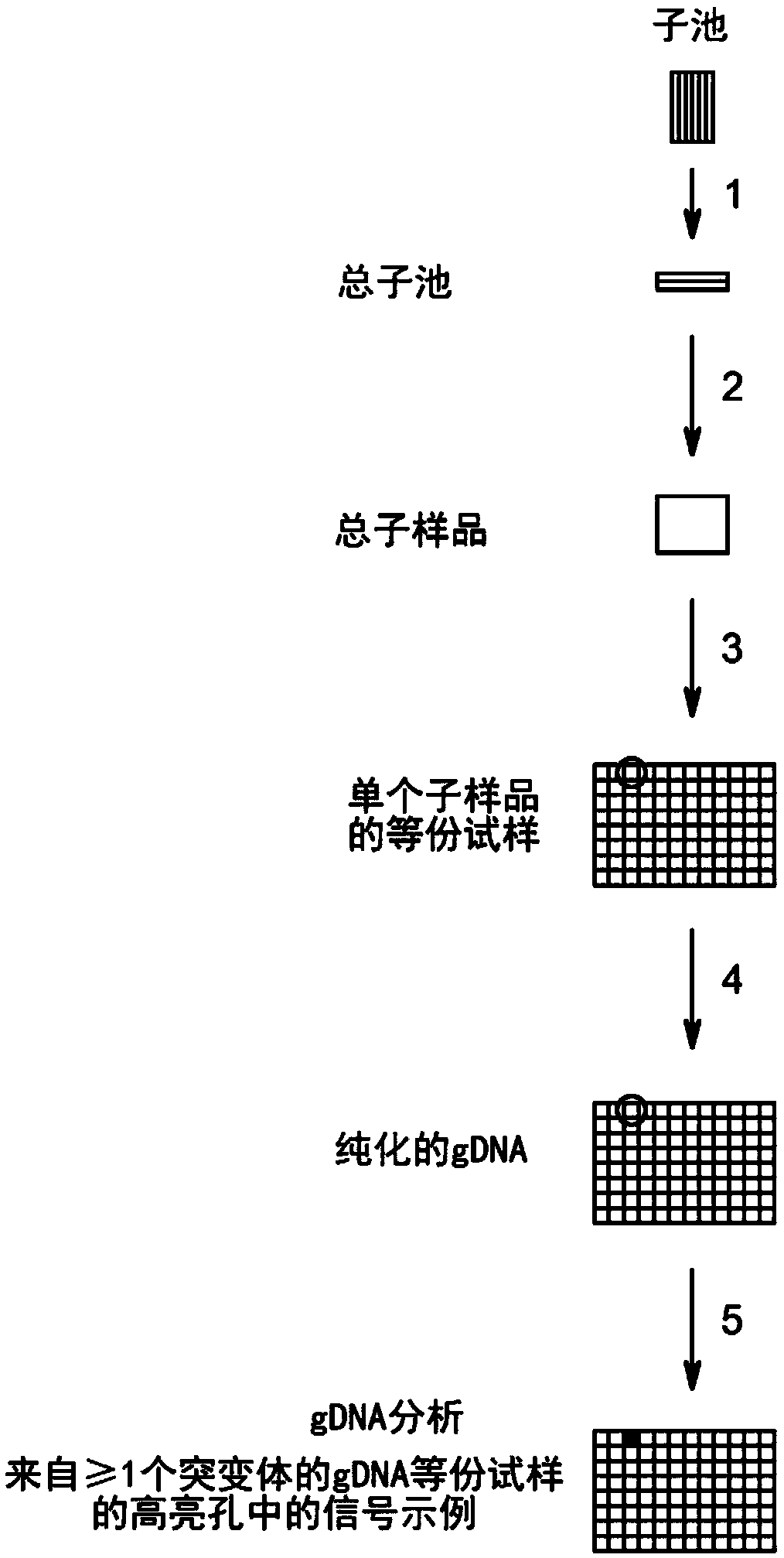
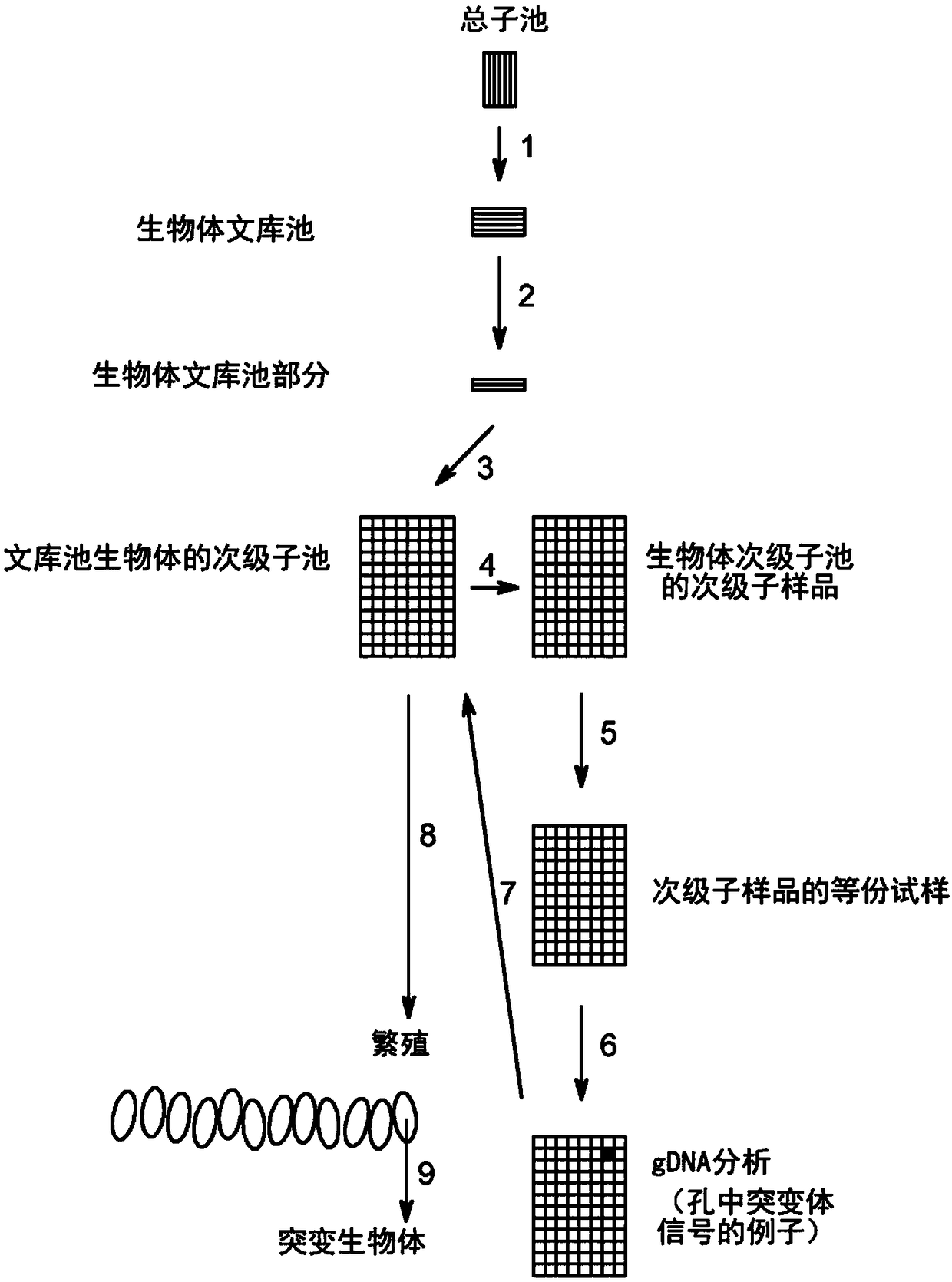
Method to screen for a mutant within a population of organisms by applying a pooling and splitting approach
In traditional plant breeding approaches, chemical mutagenesis may be utilized to introduce nucleotide substitutions at random in the genome of a plant, i.e. without possibilities to control the sitesof nucleotide changes. Because of genome complexities, the statistical probability is extremely little when it comes to finding a predetermined nucleotide substitution. The present invention, however, demonstrates how a novel, alternative use of digital polymerase chain reaction (dPCR), preferably droplet dPCR (ddPCR), is developed to exploit finding of specific nucleotide substitutions in mutated genes. The entire platform comprises a screening method with a library of mutagenized organisms, digital PCR -based systems and a set-up to propagate and analyze identified, mutated organisms.
Owner:CARLSBERG BREWERIES AS
LED-based Multi-window and multi-path detection method of digital polymerase chain reaction PCR
InactiveCN104198392AThe detection method is simpleSimple control schemeMaterial analysis by optical meansLength waveMulti path
The invention provides an LED-based multi-window and multi-path detection method of digital polymerase chain reaction PCR. The method comprises the following steps: the LEDs are adopted as detection light sources, transparent windows are arranged at the tail end of a capillary pipeline in a segmentation manner after the digital PCR finishes polymerase chain reaction; each LED with one type of wavelength is responsible for detecting one window, a type of signal possibly sent by sample drops is detected. Since the drops of the digital PCR are ordered, multiple signals in each sample drop can be detected and accurately counted. Compared with the method of detecting the same drop by adopting multiple bundles of laser, the method is simpler and more feasible, has the characteristics of simple structure and low cost, and also can be used as an inserting piece to be combined with the existing detection equipment so as to increase the working speed of daily blood detection in a hospital.
Owner:UNIV OF SCI & TECH OF CHINA
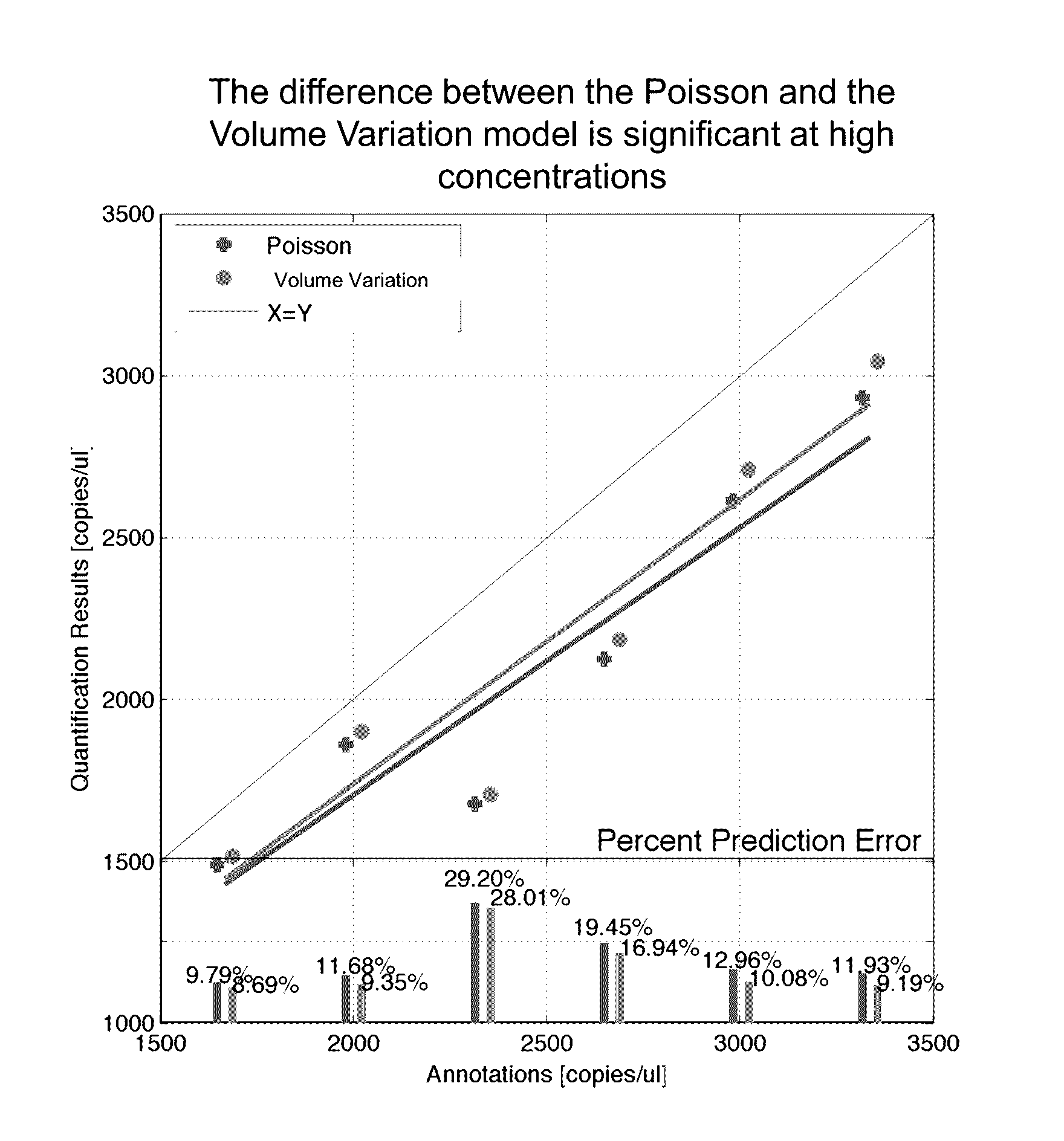
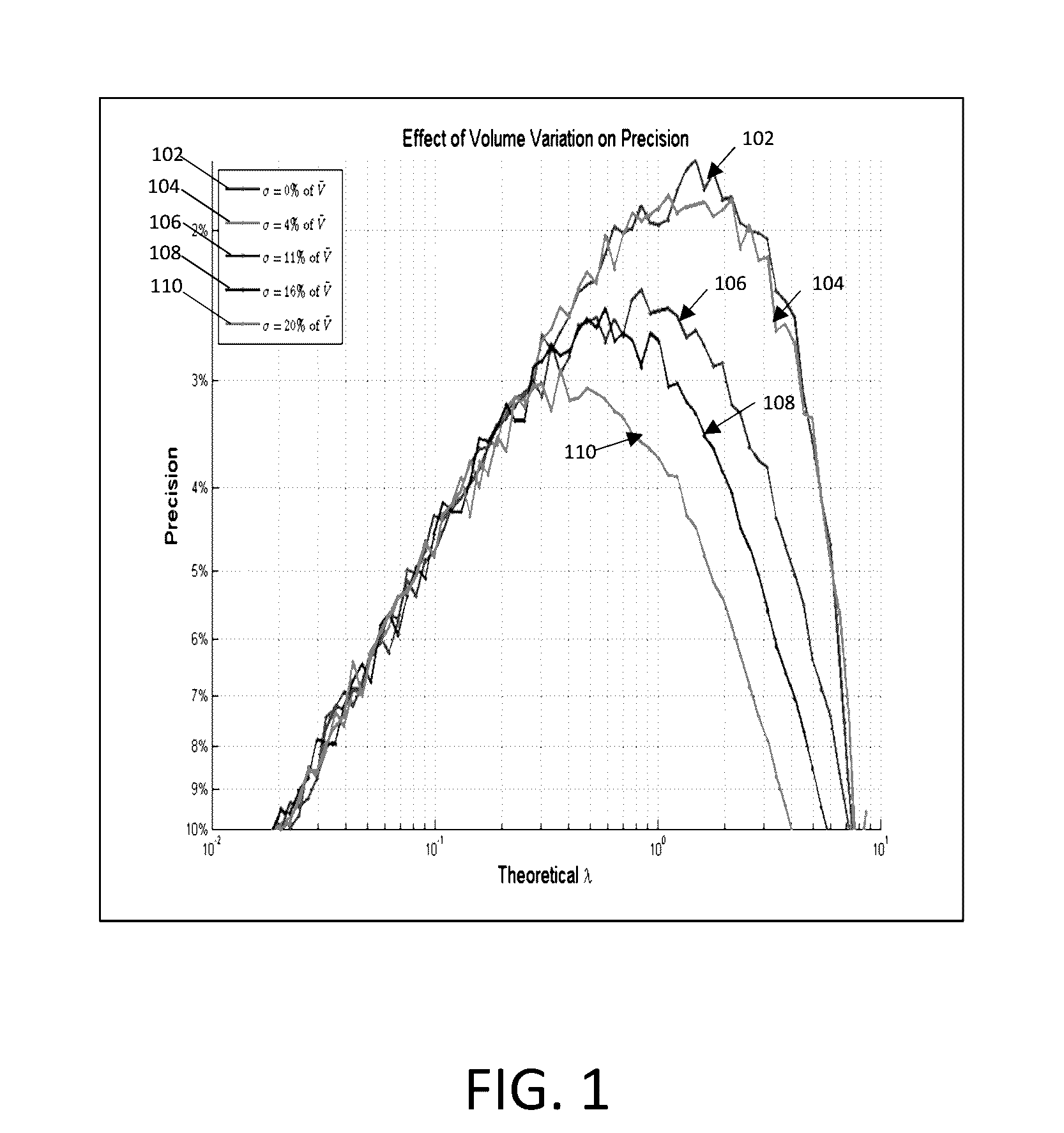
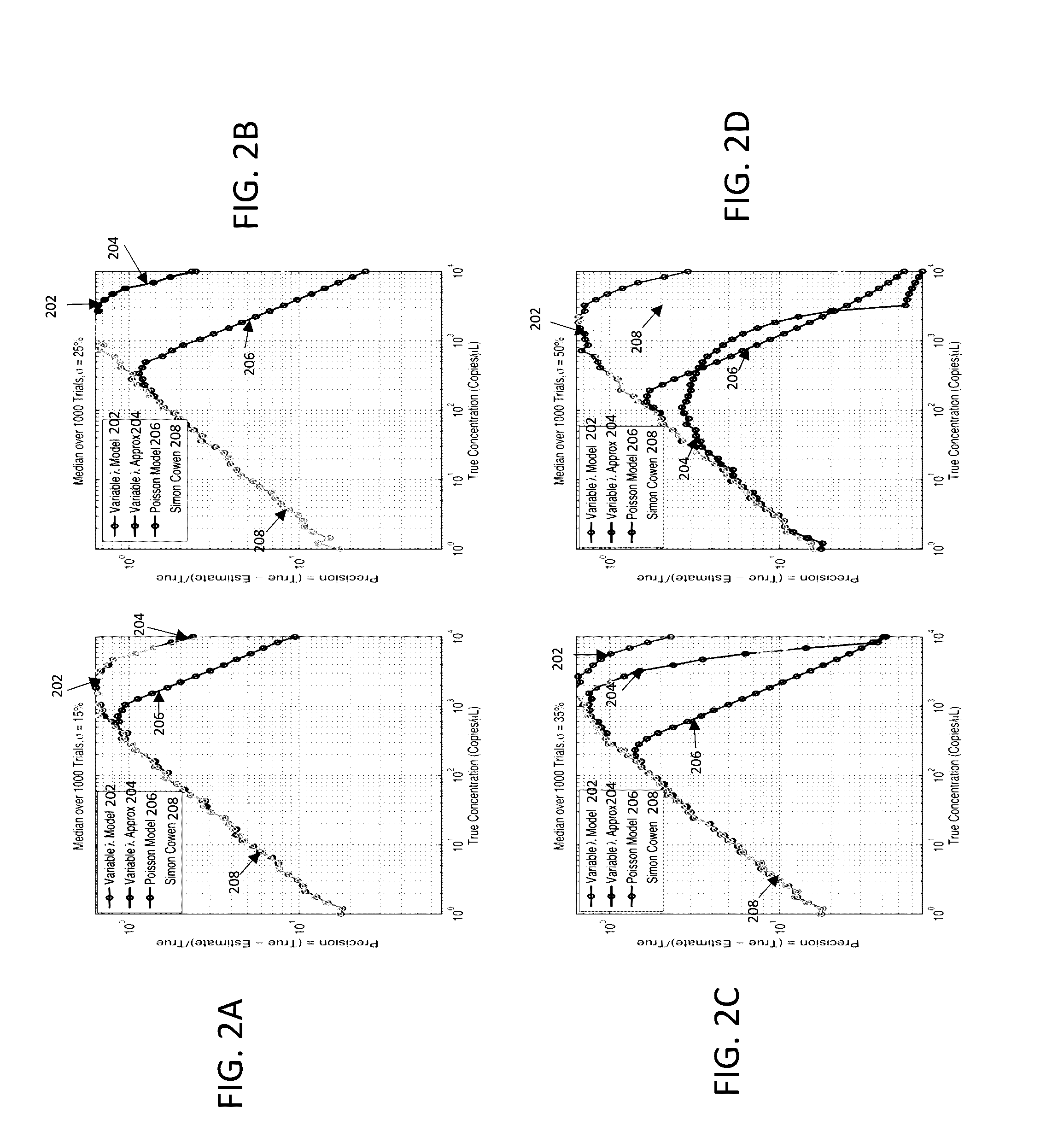
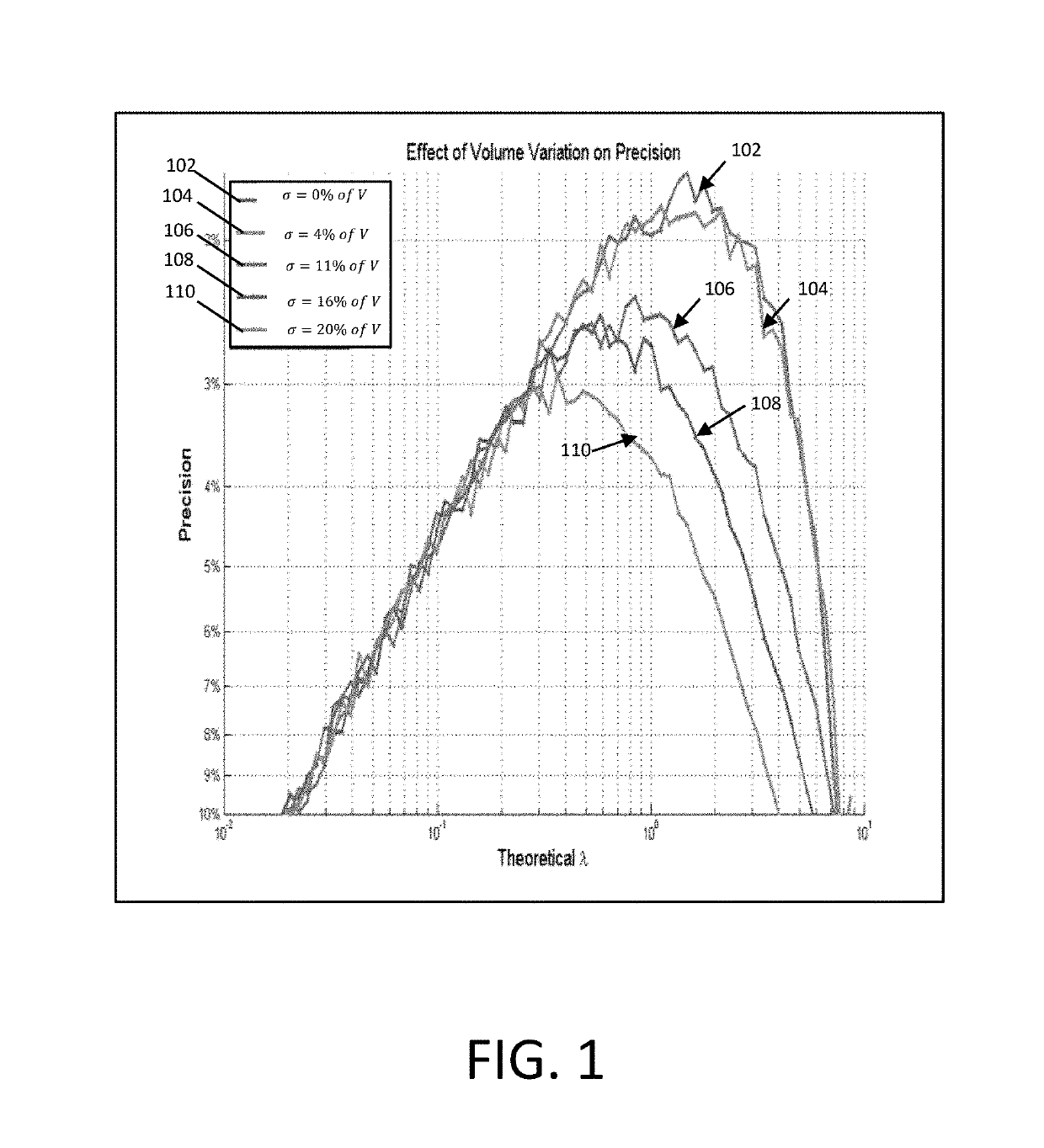
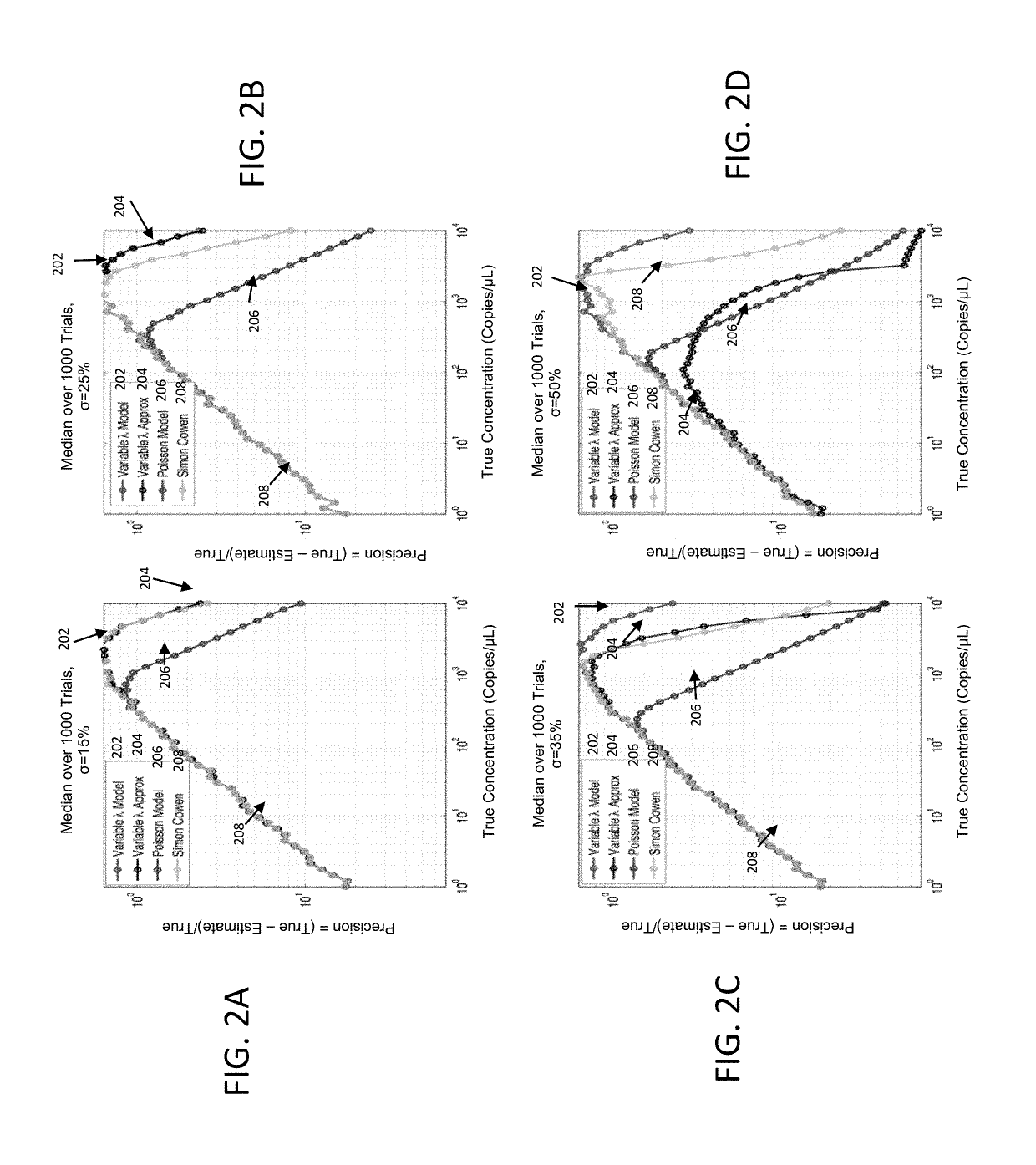
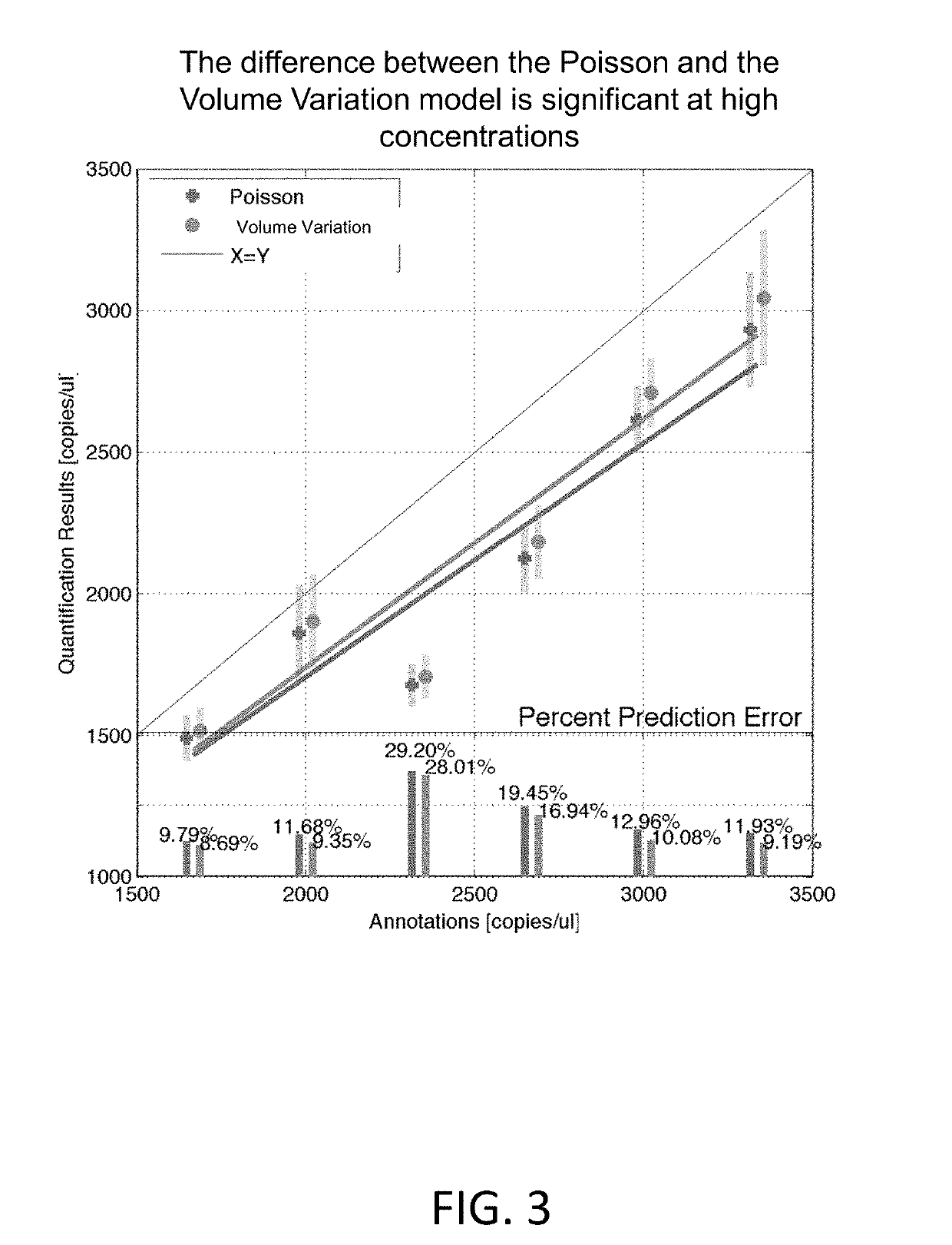
Methods and systems for volume variation modeling in digital PCR
ActiveUS20160312274A1Microbiological testing/measurementBiostatisticsVolume variationComputer science
A method for performing digital polymerase chain reaction (dPCR) is provided. The method includes partitioning a biological sample volume including a plurality of target nucleic acids into a plurality of partitions, where at least one partition includes at least one target nucleic acid. The method further includes determining a model for volume variation of the plurality of partitions and determining a number of partitions including at least one target nucleic acid. The method includes generating a concentration of target nucleic acids in the biological sample based on the model for volume variation and the fraction of partitions including at least one target nucleic acid.
Owner:LIFE TECH CORP
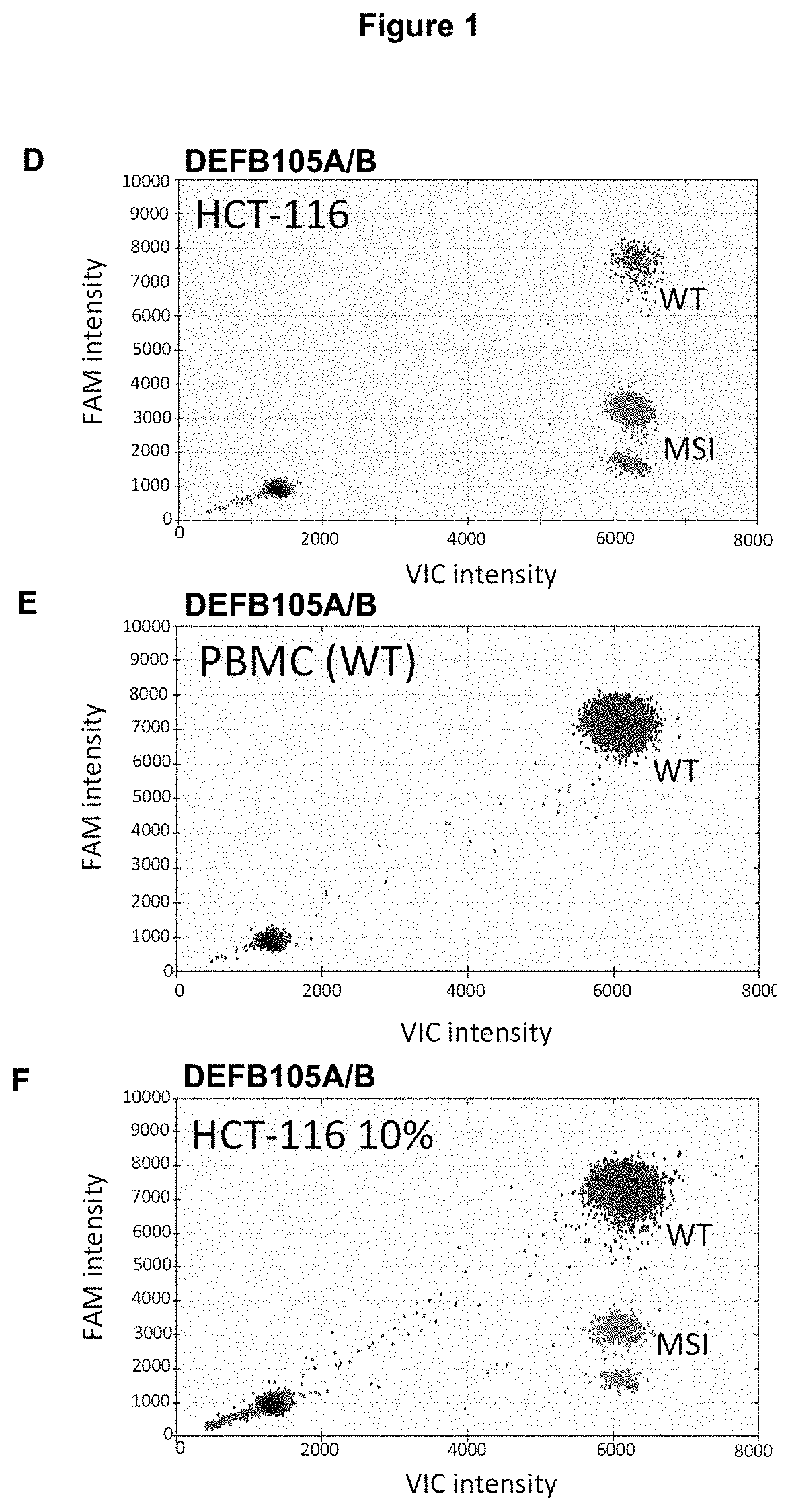
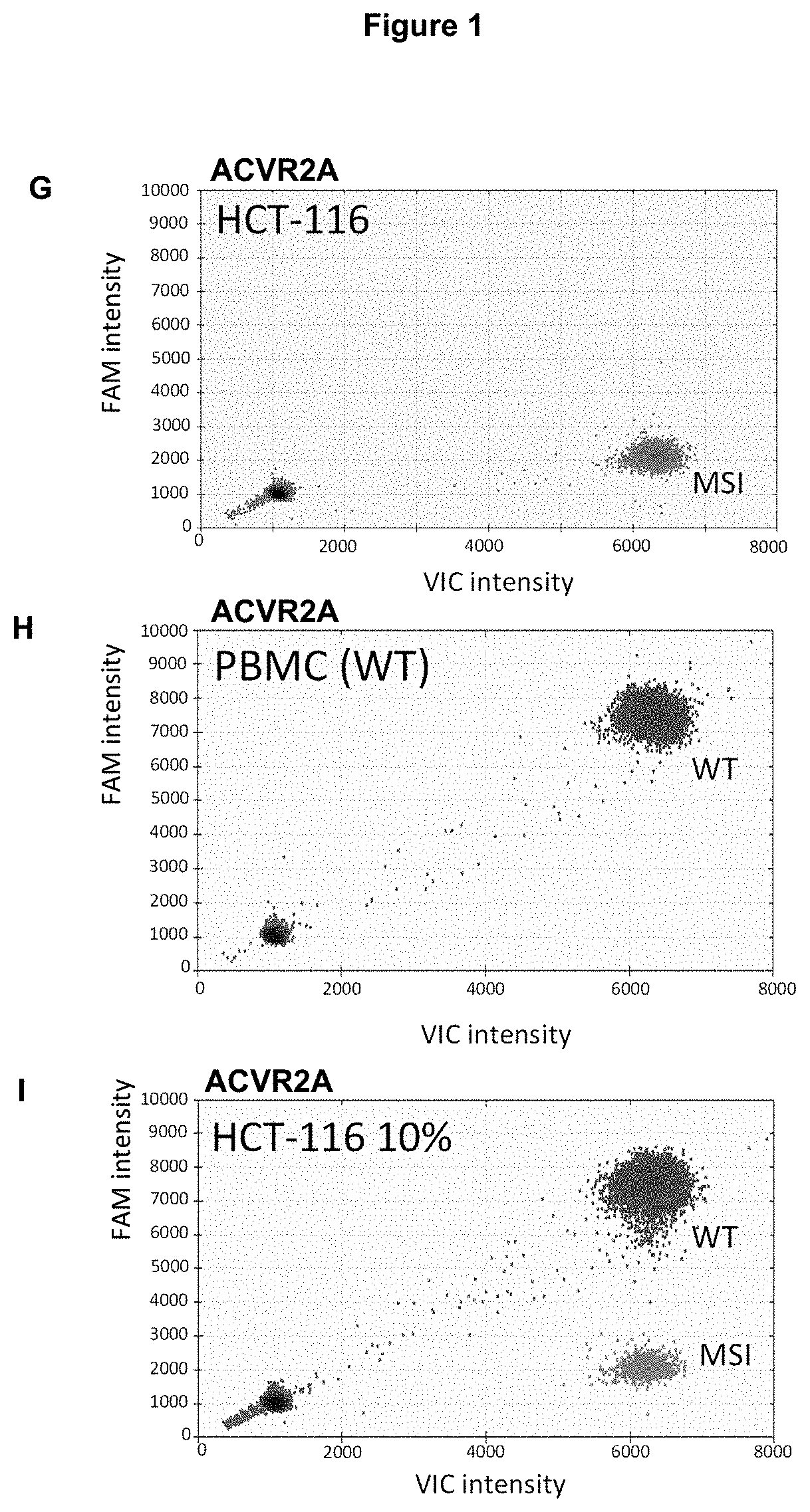
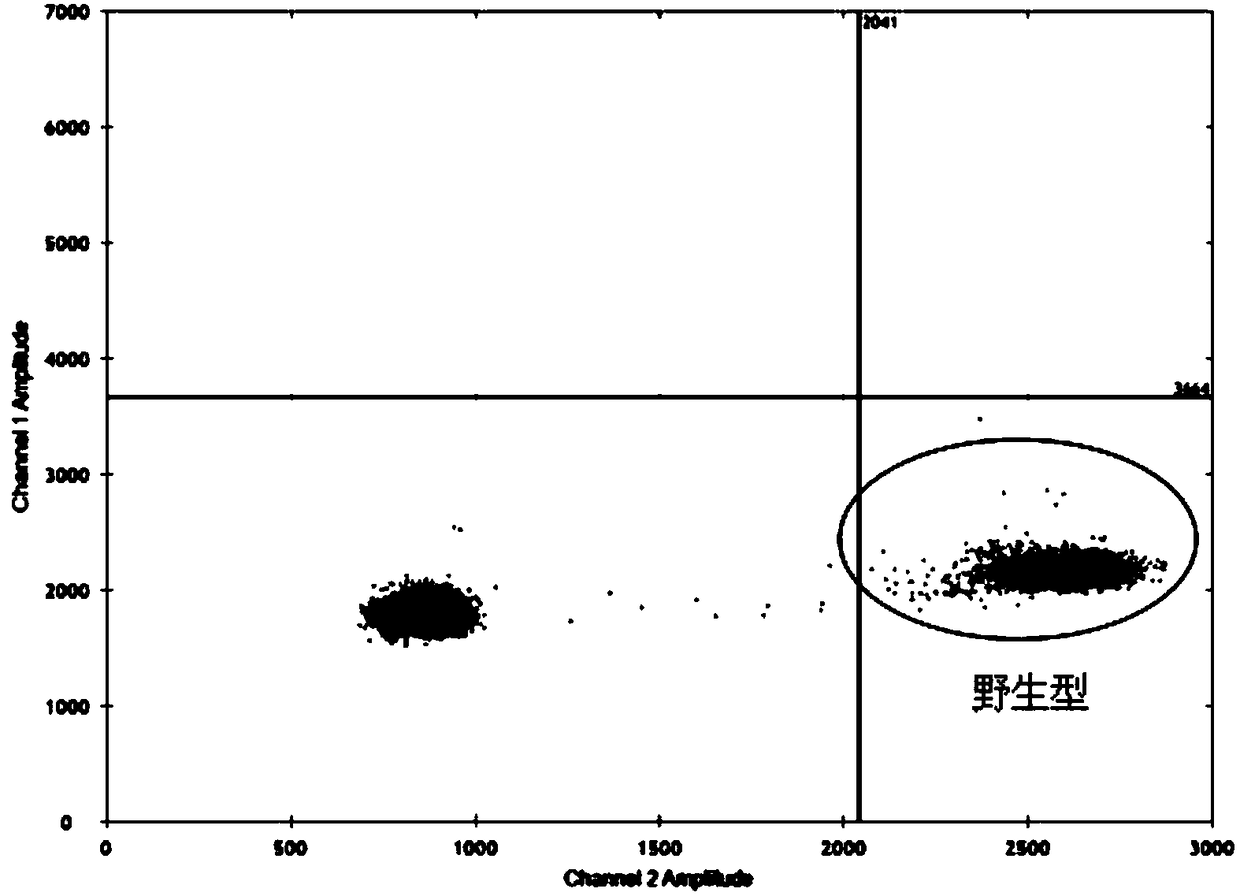
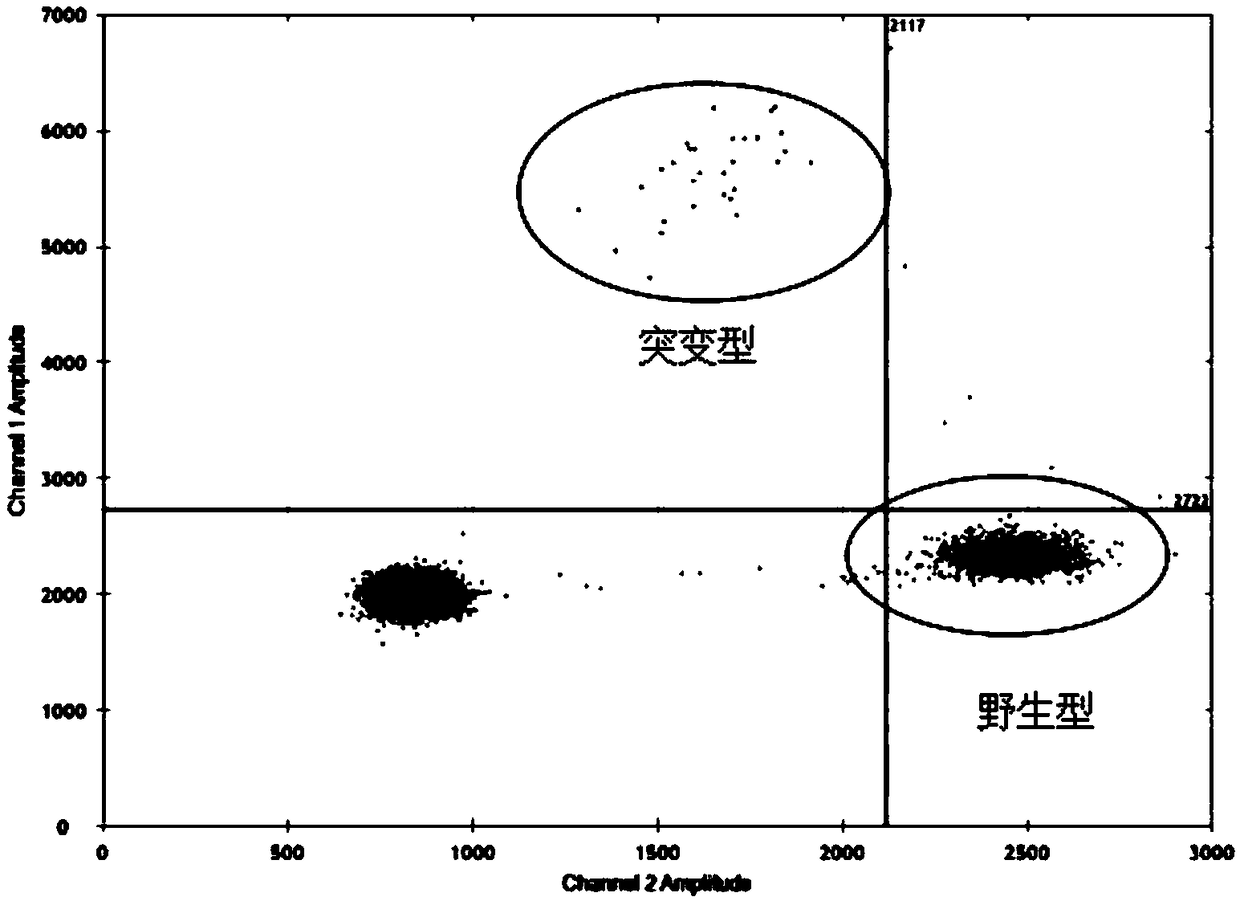
Method for Detecting a Mutation in a Microsatellite Sequence
InactiveUS20200291466A1High sensitivitySmall differenceMicrobiological testing/measurementFluorescence/phosphorescenceNucleotideMicrosatellite
The invention relates to a method for detecting a mutation in a microsatellite sequence locus of a target fragment from a DNA sample, comprising subjecting said DNA sample to a digital polymerase chain reaction (PCR) in the presence of a PCR solution comprising:a pair of primers for amplifying said target fragment of the DNA sample including said microsatellite sequence;a first MS oligonucleotide (MS) hydrolysis probe, labeled with a first fluorophore, wherein said first MS oligonucleotide probe is complementary to a wild-type sequence including the microsatellite sequence;a second oligonucleotide reference (REF) hydrolysis probe, labeled with a second fluorophore, wherein said second oligonucleotide REF probe is complementary to a wild-type sequence of said target DNA fragment which does not include said microsatellite sequence. The invention also encompasses methods for the diagnosis and prognosis of cancer and a method for determining the efficacy of a cancer treatment.
Owner:INSTITUT CURIE +2
Microfluidic system for digital polymerase chain reaction of a biological sample, and respective method
PendingCN110835600ABioreactor/fermenter combinationsBiological substance pretreatmentsBiochemistryBioinformatics
The application provides a microfluidic system for digital polymerase chain reaction (Dpcr) of a biological sample, and a respective method.
Owner:F HOFFMANN LA ROCHE & CO AG
Micro-droplet type digital polymerase chain reaction chip
InactiveCN114672412AReduce lossImprove the heating effectBioreactor/fermenter combinationsBiological substance pretreatmentsPcr chipPhysical chemistry
Owner:JIHUA LAB
Digital polymerase chain reaction detection chip, method and liquid path system thereof
ActiveCN107012079ABioreactor/fermenter combinationsBiological substance pretreatmentsEngineeringInjection device
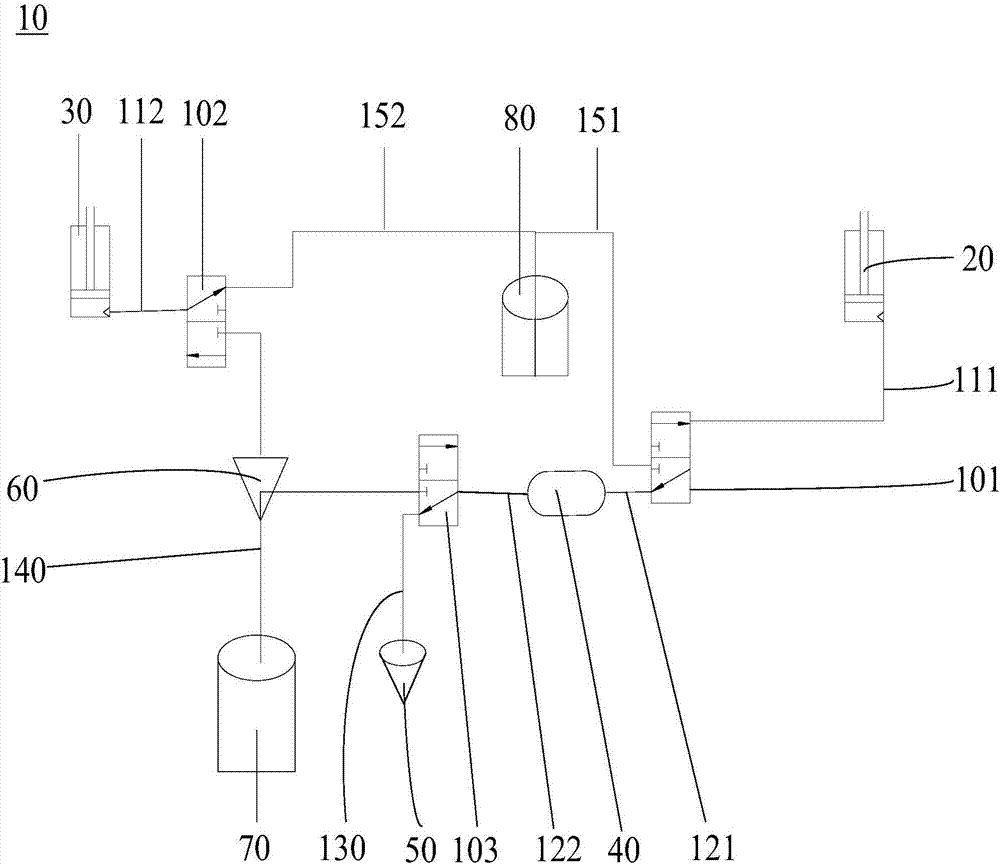
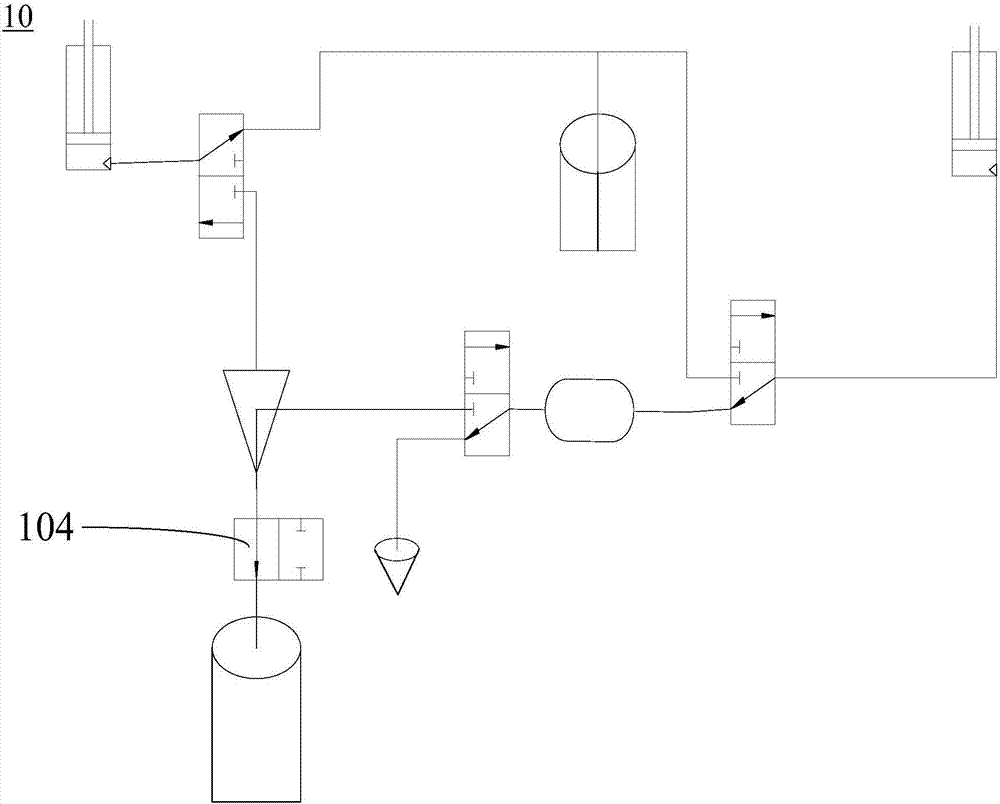
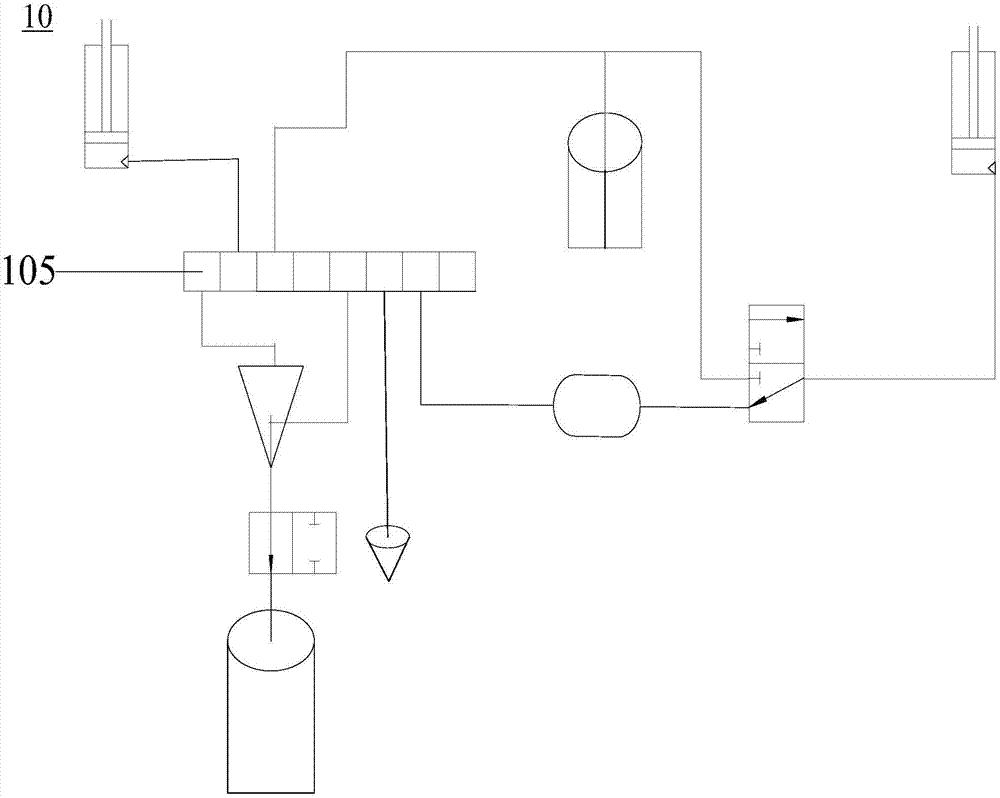
The invention relates to a digital polymerase chain reaction detection chip, a method and a liquid path system thereof. The liquid path system of the digital polymerase chain reaction detection chip comprises a first communicating valve, a second communicating valve, a third communicating valve, a first suction and injection pipeline, a second suction and injection pipeline, a first liquid storage pipeline, a second liquids storage pipeline, a sample inlet pipeline, a detection pipeline, a first oil delivery pipeline and a second oil delivery pipeline. One end of the first suction and injection pipeline is connected to the first suction and injection device while the other end is connected to the first communicating valve; one end of the second suction and injection pipeline is connected to the second suction and injection pipeline while the other end is connected to the second communicating valve; one end of the first liquid storage pipeline is connected to the first communicating valve while the other end of the first liquid storage pipeline is connected to a liquid storage device, one end of the second liquid storage pipeline is connected to the liquid storage device while the other end of the second liquid storage pipeline is connected to the third communicating valve. The pipelines of the polymerase chain reaction detection chip in the liquid path system are simple in structure and easy to clean.
Owner:广东永诺医疗科技有限公司
Compositions and methods for digital polymerase chain reaction
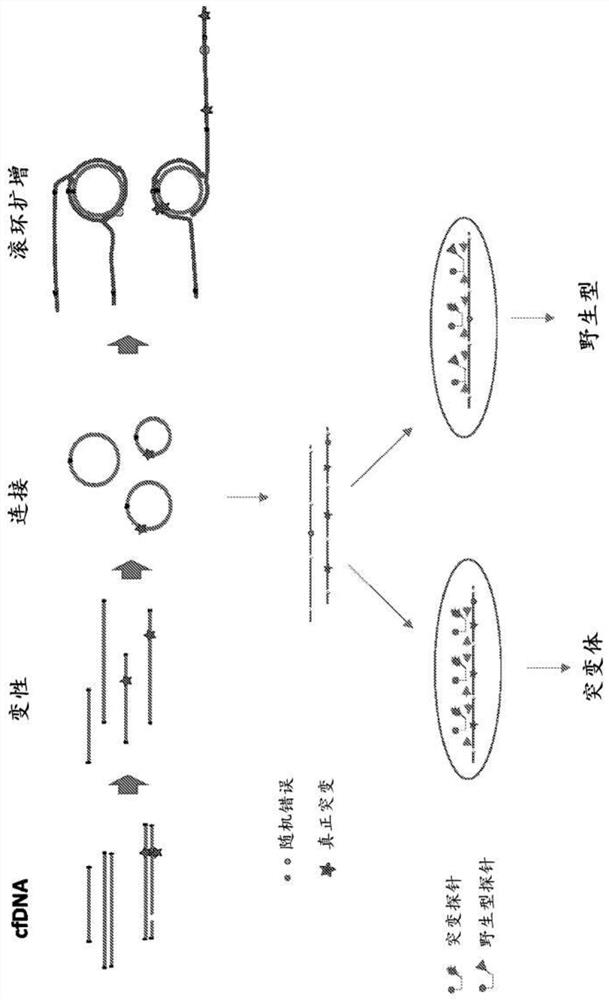
At some aspects, the present disclosure provides methods for identifying sequence variants in a nucleic acid sample. In some embodiments, a method comprises distinguishing between a true mutation in a polynucleotide and a random error introduced during an amplification step. In some embodiments, the methods reduce the number of false positives reported by a digital PCR assay. In some embodiments, the methods improve the accuracy of a digital PCR assay.
Owner:ACCURAGEN HLDG LTD
Hepatitis B virus (HBV) DNA quantitative detection kit
ActiveCN110241264ASimple designImprove the detection rateMicrobiological testing/measurementAgainst vector-borne diseasesTherapeutic effectHepatitis B virus
The invention provides a hepatitis B virus (HBV) DNA quantitative detection kit. The kit utilizes a digital polymerase chain reaction (dPCR) technology to quantitatively detect HBV DNA in biological samples and can be used for monitoring baseline level and change conditions of HBV DNA in patients suffering from hepatitis B and evaluating the response and therapeutic effect of antiviral therapy.
Owner:BEIJING DAWEI BIOTECH LTD
Digital PCR detection kit and digital PCR detection method for mutation detection of gene mutation high-incidence region
InactiveCN110863054ARich sourcesAccurate detectionMicrobiological testing/measurementDNA/RNA fragmentationConserved sequenceMutation detection
The invention discloses a digital polymerase chain reaction (PCR) detection kit and a digital PCR detection method for mutation detection of a gene mutation high-incidence region. The kit comprises apair of amplification primers, a wild-type specificity detection probe and a internal control probe; the amplification primers are used as a universal PCR amplification upstream primer and a universalPCR amplification downstream primer for detecting a plurality of mutation sites in a gene mutation high-incidence region, and an amplification region comprises a gene mutation high-incidence region to be detected; the wild-type specificity detection probe aims at the gene mutation high-incidence region; and the internal control probe aims at a gene conservative sequence in the amplification region of the gene mutation high-incidence region, wherein different fluorescent dyes are marked by the wild-type specificity detection probe and the internal control probe. The kit and the method can be used to distinguish the wild type and the mutant type of the gene mutation high-incidence region, can further determine the specific mutant type according to a positive signal position of the probes, and can realize quantification of various different mutant types to determine proportions of various mutations.
Owner:TARGETINGONE CORP +1
Method for detecting ARMS-ddPCR (amplification refractory mutation system-droplet digital polymerase chain reaction) gene site-directed mutation
InactiveCN109593832AImproved prognosisLow costMicrobiological testing/measurementSmall fragmentWild type
The invention discloses a method for detecting ARMS-ddPCR (amplification refractory mutation system-droplet digital polymerase chain reaction) gene site-directed mutation. The method aims at gene mutation of one site, primers comprise a wild type primer and a mutation primer, probes comprise a wild type probe and at least one mutation probe, a mutation site is detected by further combination witha ddPCR platform, and sample mutation site DNA is subjected to absolute quantification. The method is particularly suitable for amplification of small-fragment DNA samples such as plasma free DNA (cfDNA) and the like, with application of the method, gene mutation with quite low abundance can be detected from cfDNA, the method has the characteristics of good specificity and high sensitivity, can beapplied to absolute quantification detection of the gene mutation site by the ddPCR platform and has a quite important effect on guidance of clinic treatment and improvement of patient outcome.
Owner:江苏苏博生物医学科技南京有限公司 +1
Nucleic acid combination for detecting pseudorabies virus and application of nucleic acid combination, and kit and method for detecting pseudorabies virus
PendingCN106636460ARealize detectionAvoid missing detectionMicrobiological testing/measurementMicroorganism based processesConserved sequenceVirus detection
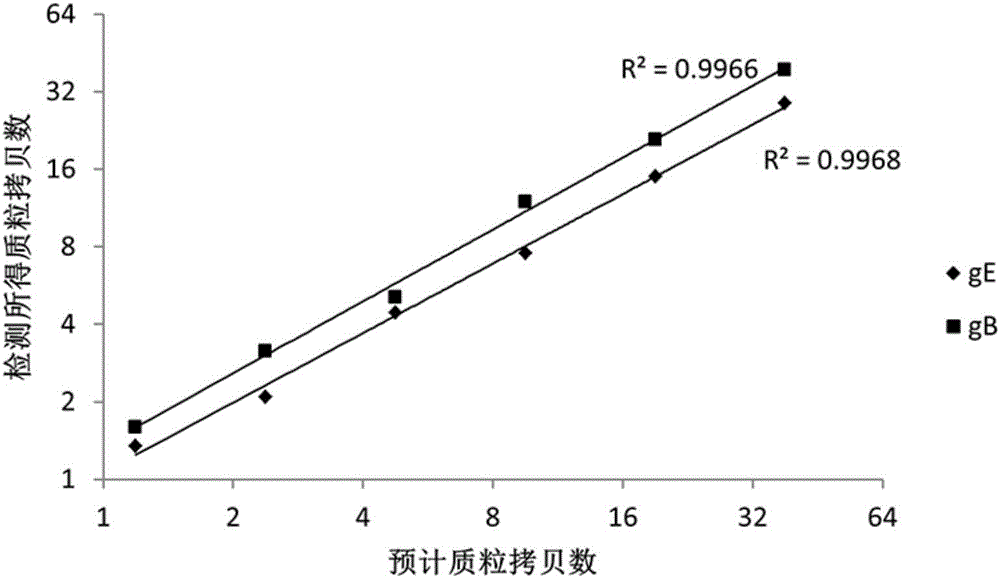
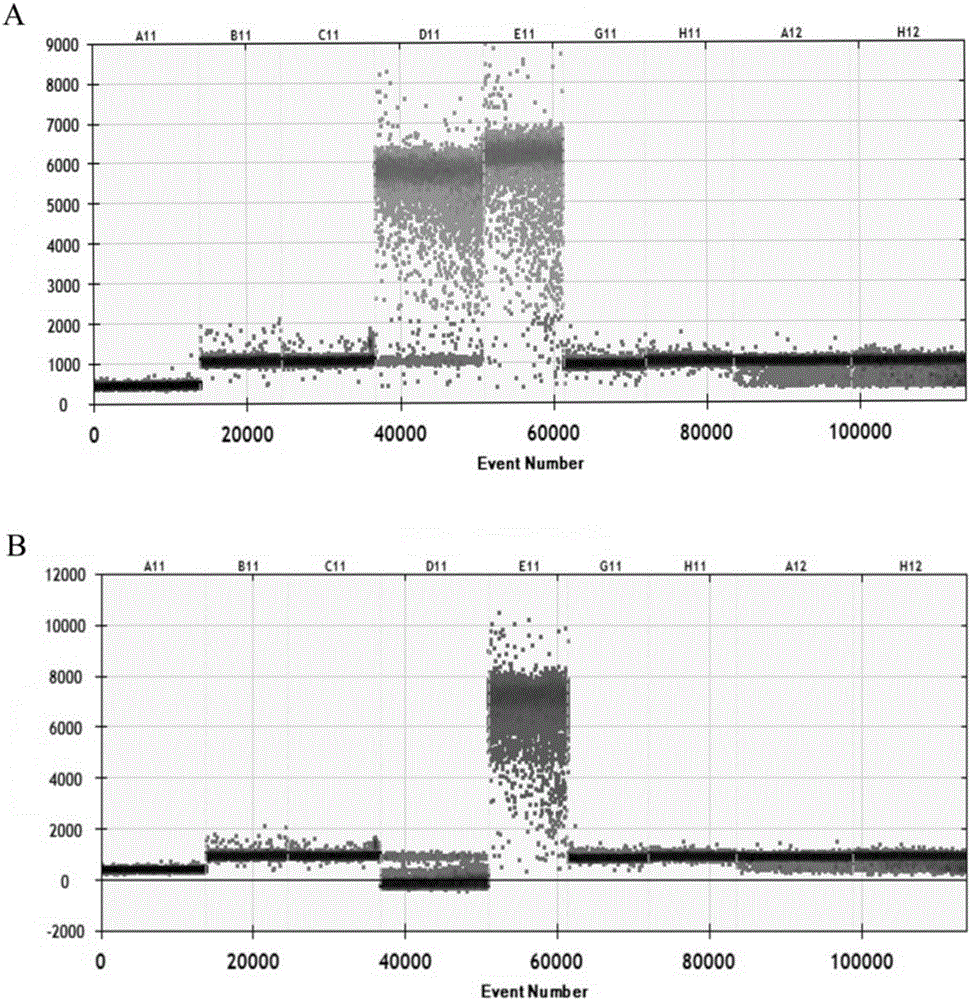
The invention provides a nucleic acid combination for detecting pseudorabies virus and application of the nucleic acid combination, and a kit and a method for detecting the pseudorabies virus, and relates to the technical field of pseudorabies virus detection. The nucleic acid combination for detecting the pseudorabies virus comprises a first primer pair designed according to a conserved sequence of a gB gene of the pseudorabies virus, and a first probe, wherein the first primer pair comprises two primers, and the base sequences of the two primers are respectively as shown in SEQ ID No. 1 and SEQ ID No. 2; the base sequence of the first probe is as shown in SEQ ID No. 3. After the nucleic acid combination is adopted, rabies virus can be rapidly and conveniently detected on a digital polymerase chain reaction (PCR) platform, samples with low rabies virus content can be detected, and missed detection is avoided; the nucleic acid combination has the characteristics of being high in sensitivity, high in specificity, high in detection rate, and the like.
Owner:成都海关技术中心 +1
Circulating digital PCR method, circulation system, digital PCR chip and preparation method of digital PCR chip
ActiveCN110804650AEasy to operateReduce gas residueBioreactor/fermenter combinationsBiological substance pretreatmentsPcr chipTemperature control
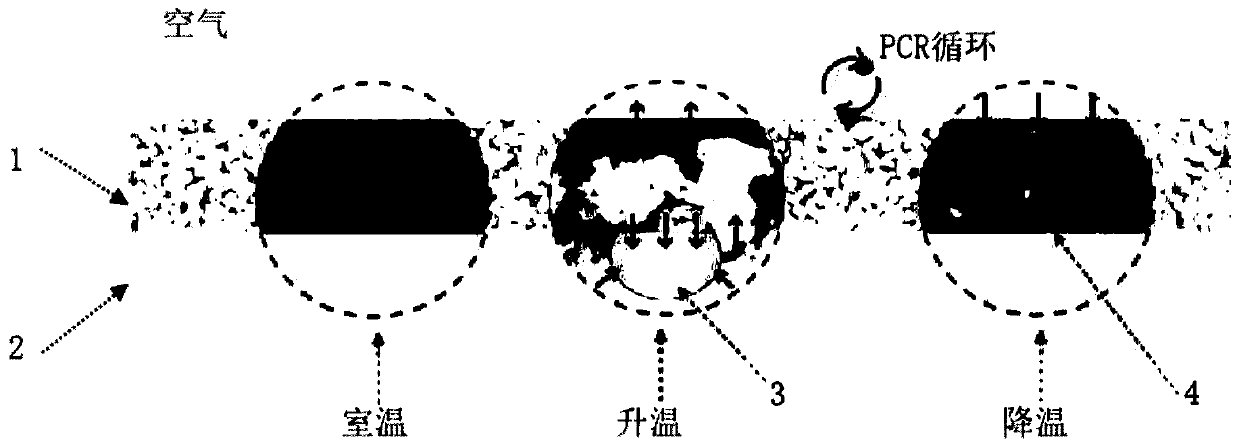
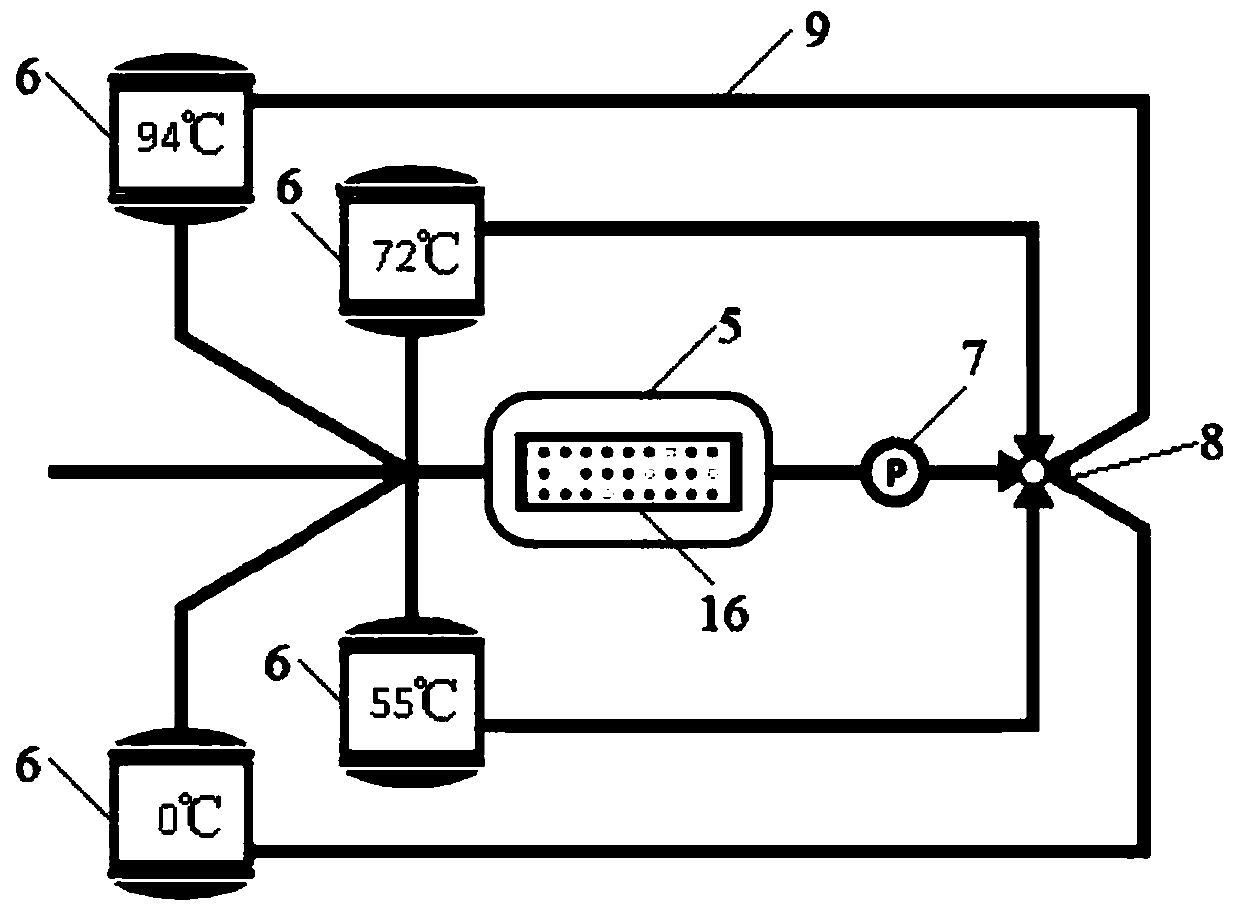
The invention discloses a circulating digital polymerase chain reaction (PCR) method, a circulation system, a digital PCR chip and a preparation method of the digital PCR chip. The digital PCR methodincludes the following steps: S1, placing a digital PCR chip subjected to injection processing into a reaction chamber of the circulation system; S2, draining air in the reaction chamber, and pressurizing the circulation system; S3, starting the circulation system for a PCR reaction; S4, depressurizing the circulation system after the PCR reaction is completed; and S5, taking out the digital PCR chip in the reaction chamber, and performing fluorescence signal analysis on the digital PCR chip. The sampling of the chip provided by the invention does not rely on complex equipment such as pumps and valves, and does not need to use high-viscosity thermal polymerization separation oil, the chip does not need to be sealed after the injection is completed, so that the operation is simple; the chiphas a small thickness, fast heat conduction, and a rapid reaction; a constant-temperature liquid storage tank has a large volume and rapid heat exchange, can quickly heat or cool the chip, and has good temperature control, and no temperature overshoot; and the chip has a simple structure and low costs.
Owner:SHANGHAI INST OF MICROSYSTEM & INFORMATION TECH CHINESE ACAD OF SCI
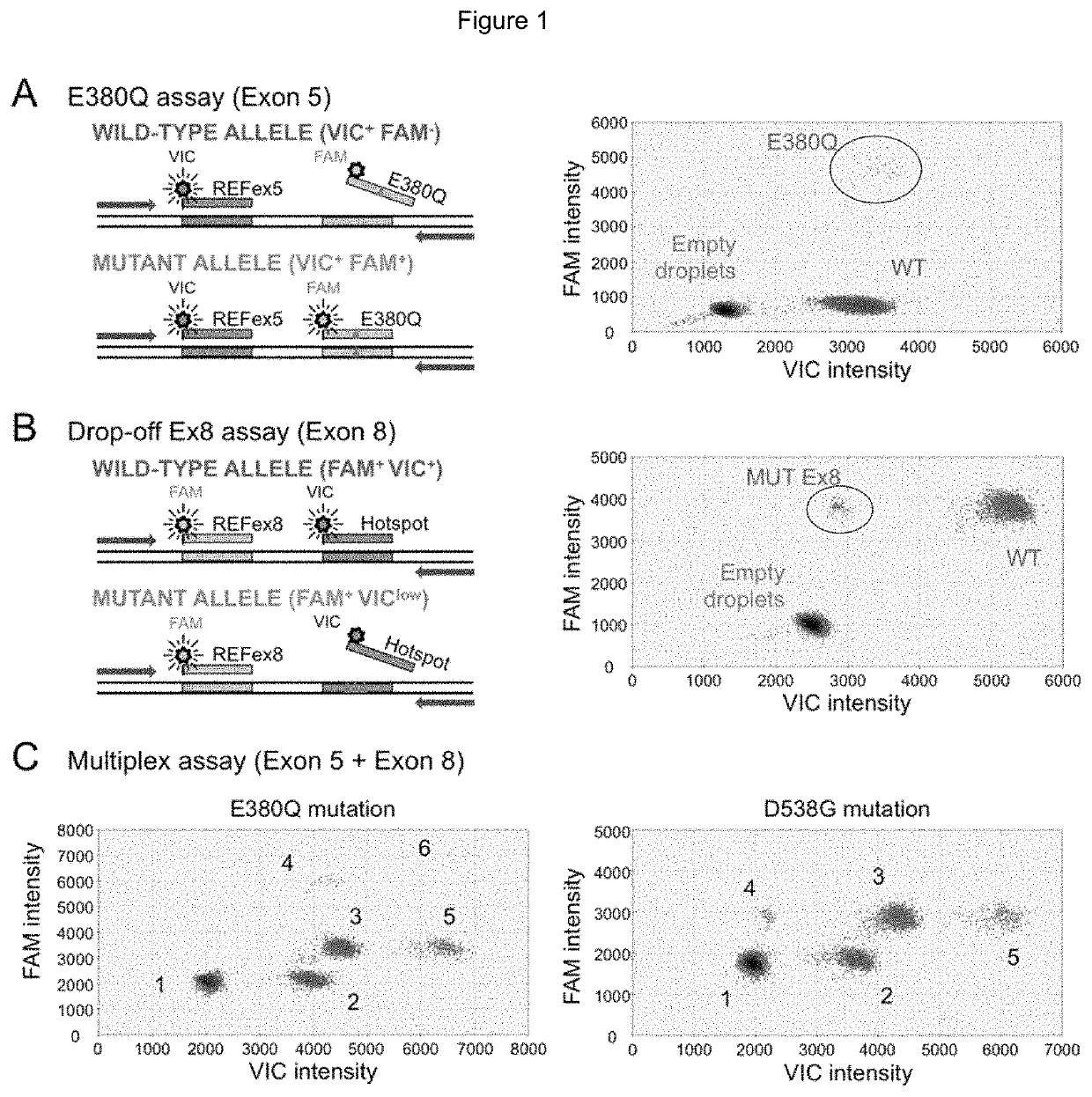
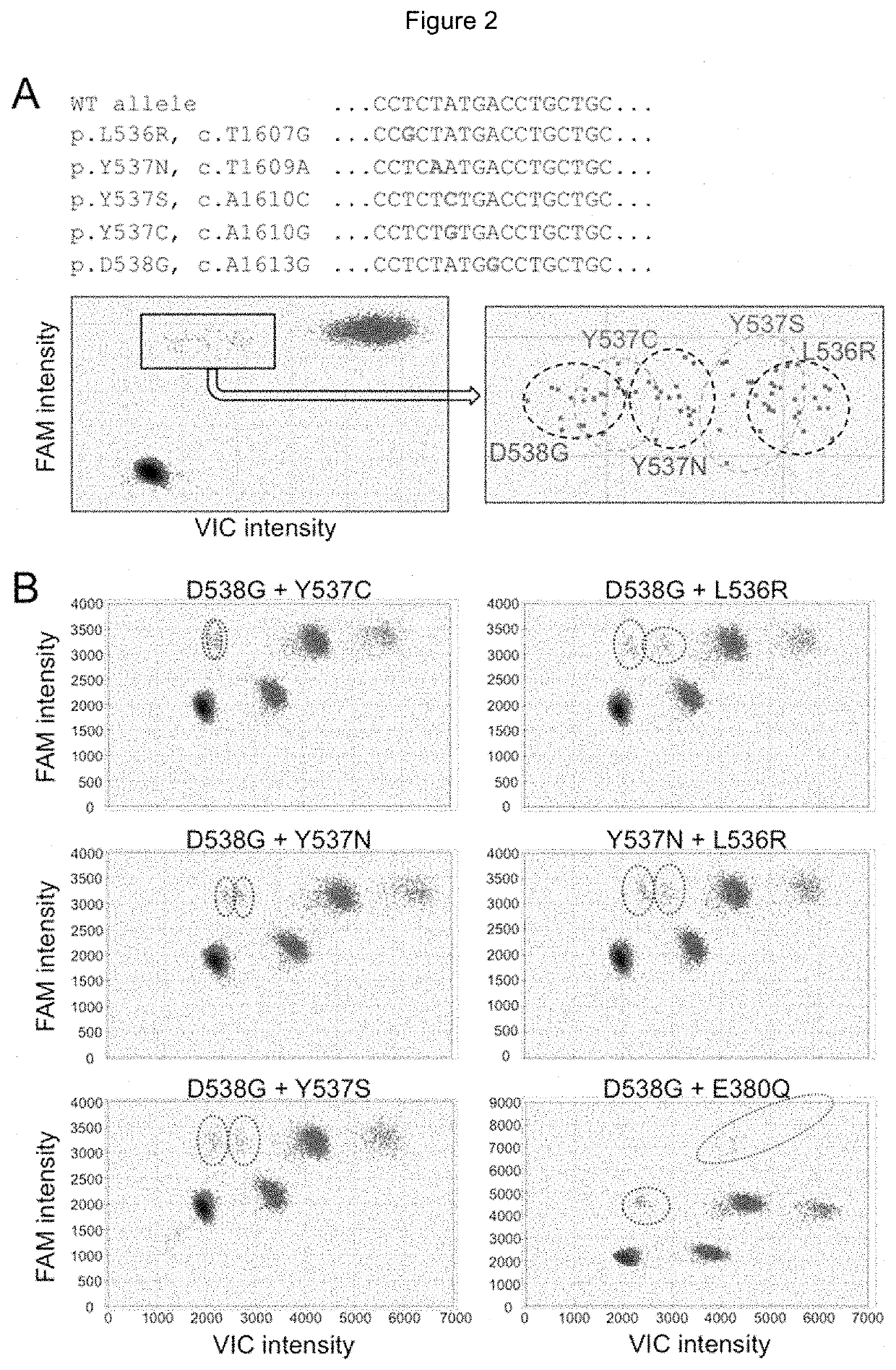
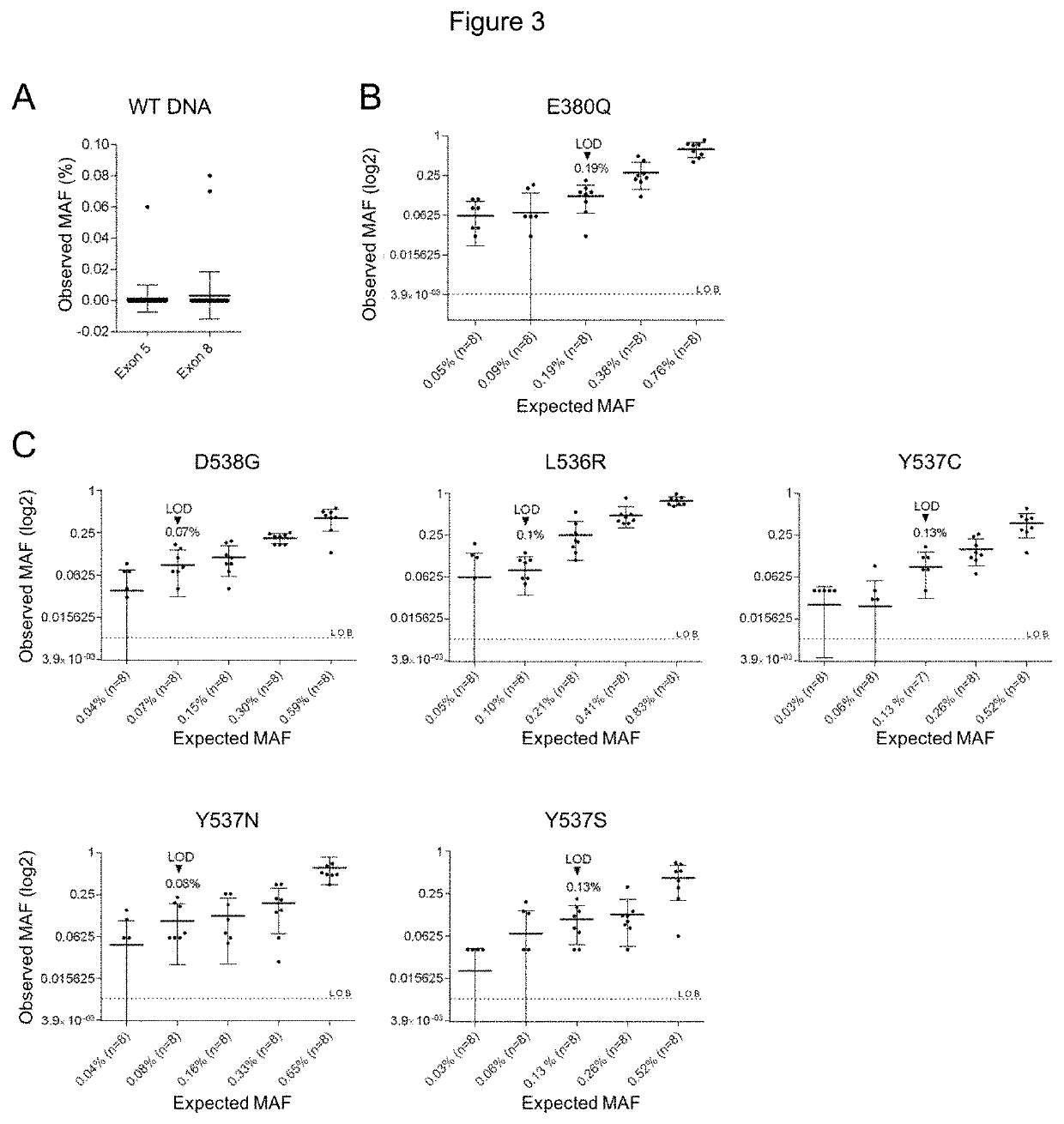
Method for Identifying One or More Mutations in a Hotspot Mutation Sequence
The present invention relates to an in vitro method for identifying and / or characterizing one or more mutations in a hotspot mutation sequence of at least one ESR1 target fragment from a DNA sample,said method comprising subjecting the DNA sample to a drop-off digital polymerase chain reaction (PCR) in the presence of a PCR solution comprising:a pair of primers suitable for amplifying an ESR1 target fragment;an oligonucleotide reference (REF) hydrolysis probe, labeled with a fluorophore, wherein said REF oligonucleotide probe is complementary to a wild-type sequence of the target fragment located outside of the hotspot mutation sequence;an oligonucleotide hotspot (HOTSPOT) hydrolysis probe, labeled with another fluorophore, wherein said oligonucleotide HOTSPOT probe is complementary to a wild-type sequence of the hotspot mutation sequence of the target DNA fragment.
Owner:UNIVERSITÉ PARIS CITÉ
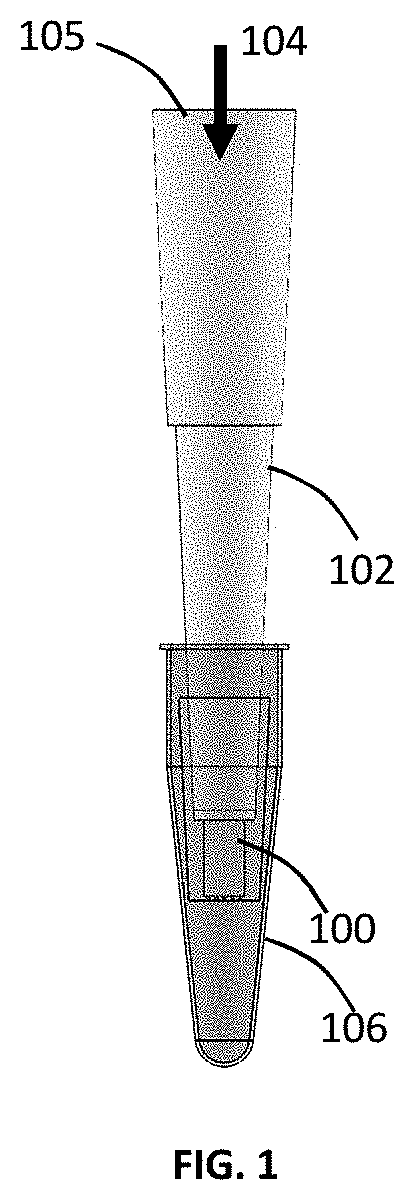
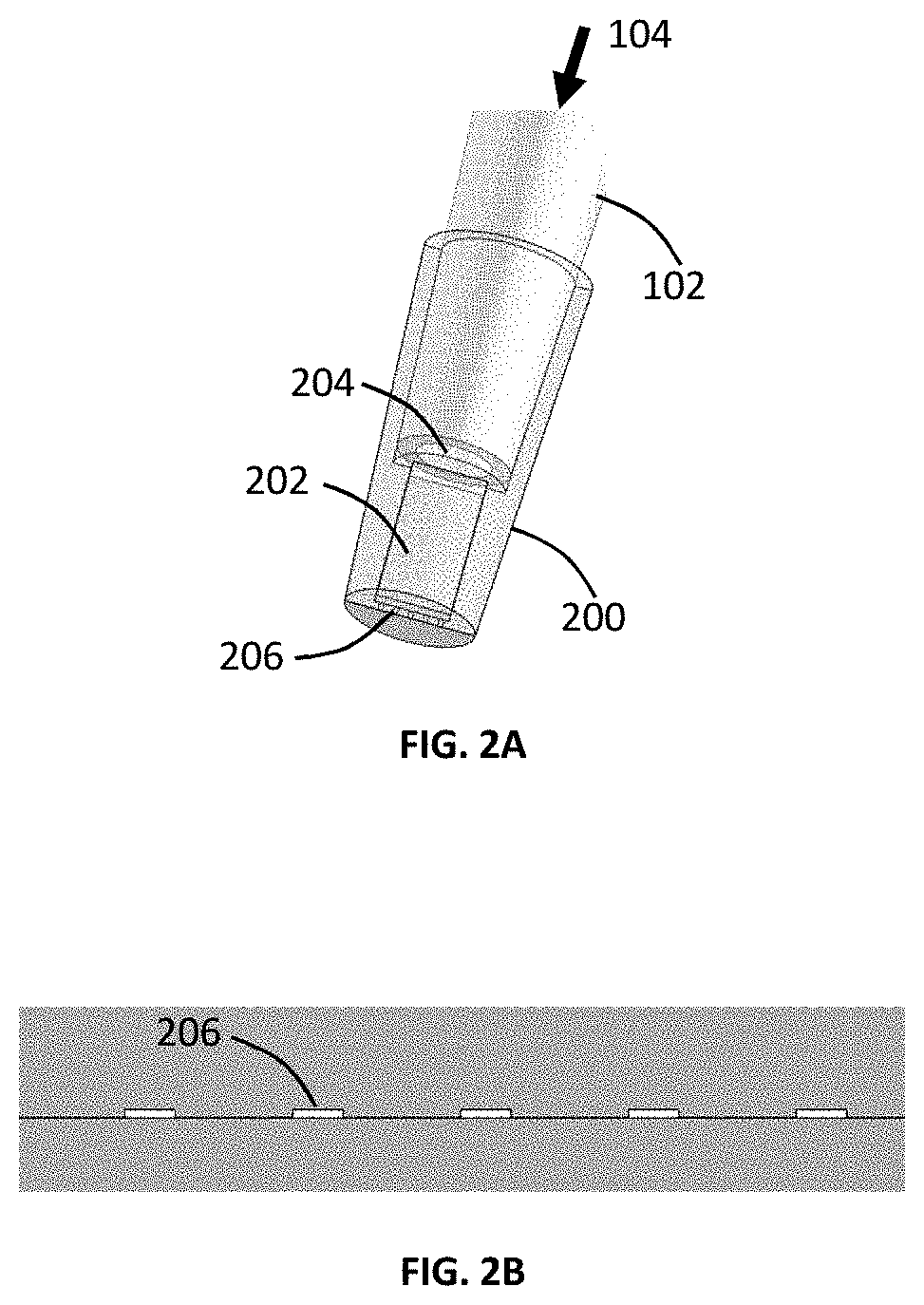
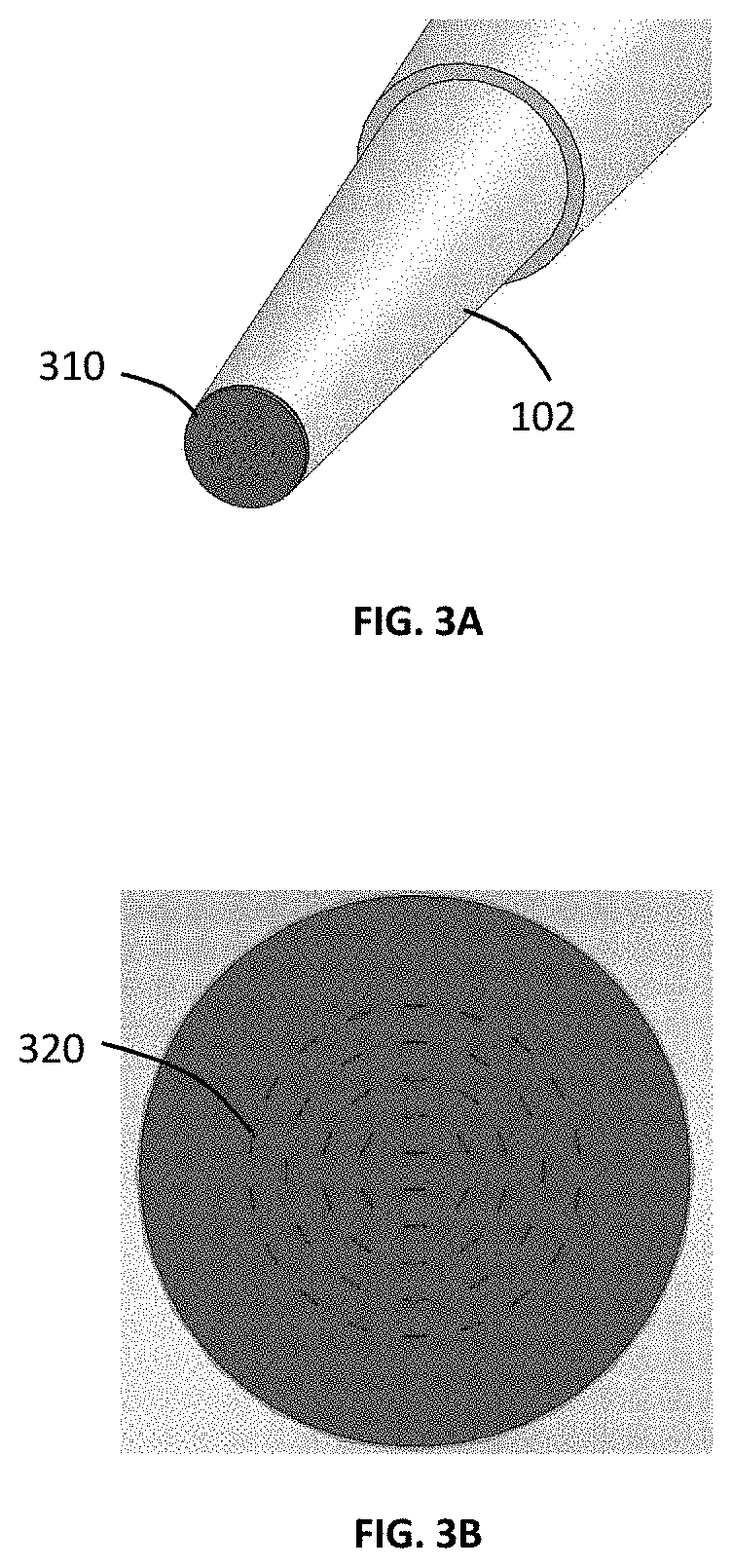
Micro-pipette tip for forming micro-droplets
ActiveUS20200101454A1Low costGenerate flexibleFlow mixersTransportation and packagingFluid phasePolymerase
A micro-droplet-emulsifier to generate a micro-droplet-emulsion for application on digital polymerase chain reaction is provided. This device comprises of a micro-pipette to hold a dispersed-phase-liquid; a droplet generator that attaches to the micro-pipette and has a plurality of substantially flat micro-channels. The dispersed-phase-liquid is forced through the micro-channels to form micro-droplet-emulsion of dispersed-liquid-phase in the continuous-liquid-phase inside the chamber. The size of the micro-droplets is controlled by the shape and the aspect ratio of the micro-channels, the depth of micro-channel and the material of the micro-droplet-generator-head that dictates the contact angle of the droplet on the micro-channels.
Owner:MOLARRAY RES INC
Methods and systems for visualizing and evaluating data
ActiveCN104093853AMicrobiological testing/measurementData visualisationComputer scienceEvaluated data
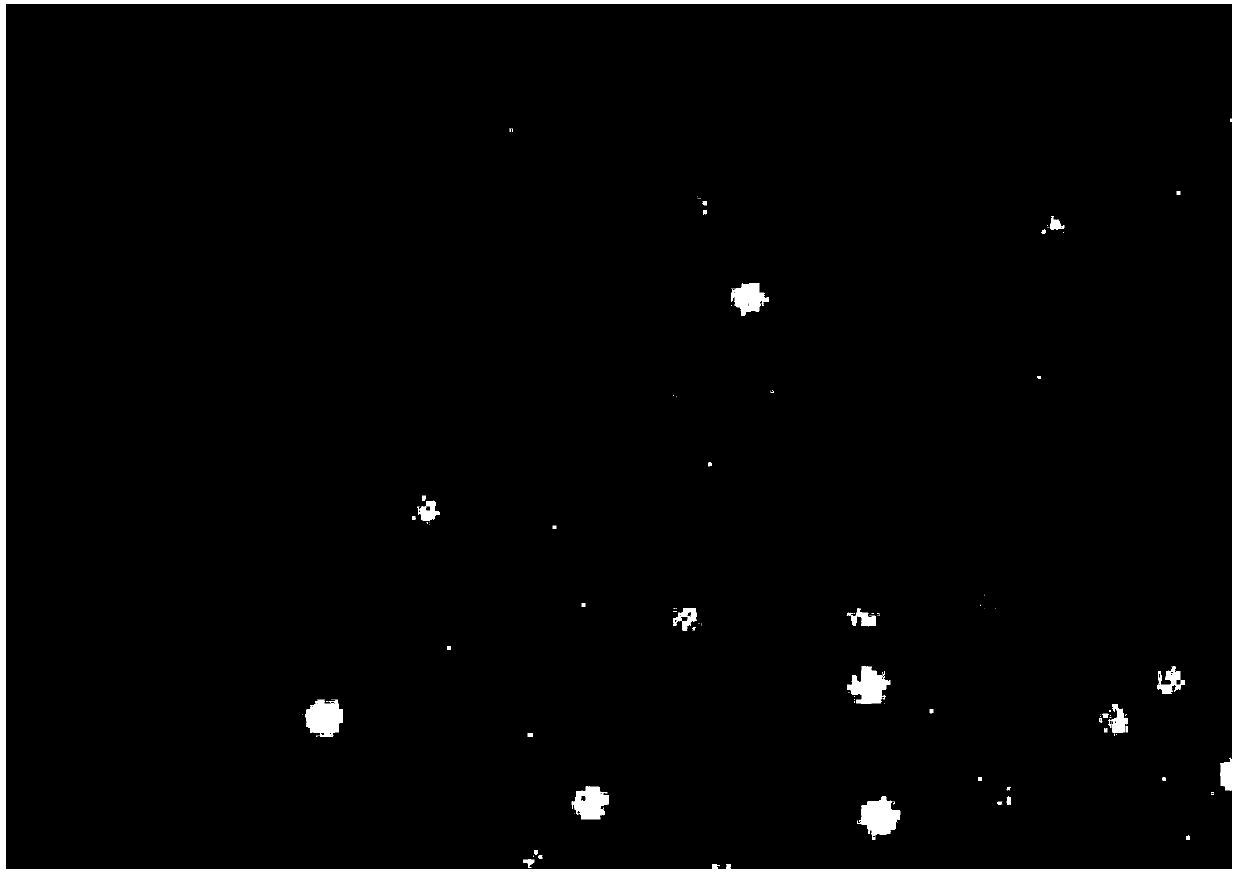
A computer-implemented method of generating a digital polymerase chain reaction (dPCR) result is provided. The method includes detecting a first set of emission data from a plurality of samples, each included in a sample region of a plurality of sample regions, at a first time during an amplification period. The method further includes determining a positive or negative amplification determination for each sample of the plurality of samples based in part on the first set of emission data. A dPCR result is generated based on the positive or negative amplification determinations for the plurality of samples.
Owner:LIFE TECH CORP
Primers, reagent kit and method for detecting IDH1 (isocitrate dehydrogenase 1) R132H gene variation by aid of ddPCR (droplet digital polymerase chain reaction) technologies
InactiveCN108384857AImproved prognosisStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationIDH1Small fragment
The invention discloses primers, a reagent kit and a method for detecting IDH1 (isocitrate dehydrogenase 1) R132H gene variation by the aid of ddPCR (droplet digital polymerase chain reaction) technologies. The method includes extracting cfDNA (cell-free deoxyribonucleic acid) in peripheral blood; designing two probes and a pair of primers for IDH1R132H mutant types and wild types; carrying out ddPCR amplification by the aid of the cfDNA used as a template; carrying out data analysis on obtained amplification products to obtain absolute quantities and proportions of IDH1R132H mutant genes. Thereagent kit for detecting the IDH1R132H gene variation is based on the method. The primers, the reagent kit and the method have the advantages that provided primer amplification fragments 83bp are particularly applicable to amplifying small-fragment DNA samples such as plasma free DNA and can be combined with the detection probes, accordingly, the IDH1R132H gene variation with extremely low abundance can be detected from the cfDNA, the primers, the reagent kit and the method are good in specificity and high in sensitivity, human IDH1R132H gene variation can be absolutely quantitatively detected by the aid of the primers, the reagent kit and the method, and important effects can be realized by the primers, the reagent kit and the method for guiding clinical treatment and improving patientprognosis; the IDH1R132H gene variation can be detected only by the aid of the peripheral blood without tissue specimen collection, and accordingly the method is particularly suitable for patients intolerant to tissue sampling.
Owner:PRIMBIO GENES BIOTECH WUHAN CO LTD
High-precision specific digital polymerase chain reaction (PCR) detection method for series infectious microbe contamination taking acetobacter aceti as priority in fermented milk
PendingCN110592195AMicrobiological testing/measurementMicroorganism based processesBiotechnologyAcetobacter aceti
The invention discloses a high-precision specific digital polymerase chain reaction (PCR) detection method for series infectious microbe contamination taking acetobacter aceti as priority in fermentedmilk. The method comprises the steps: firstly, to-be-detected infectious microbes are subjected to dead bacterium de-interference treatment, then bacterial DNA is subjected to low-impurity and high-purity extraction, a specific primer probe of the to-be-detected microbes is researched, developed and designed, a ddPCR system and procedure and the annealing temperature thereof are set, and thus thedetection method meets the requirements of specificity, sensitivity and quantification. The constructed method is good in specificity, has no non-specific amplification, and has good sensitivity andquantitative advantages.
Owner:河北省食品检验研究院
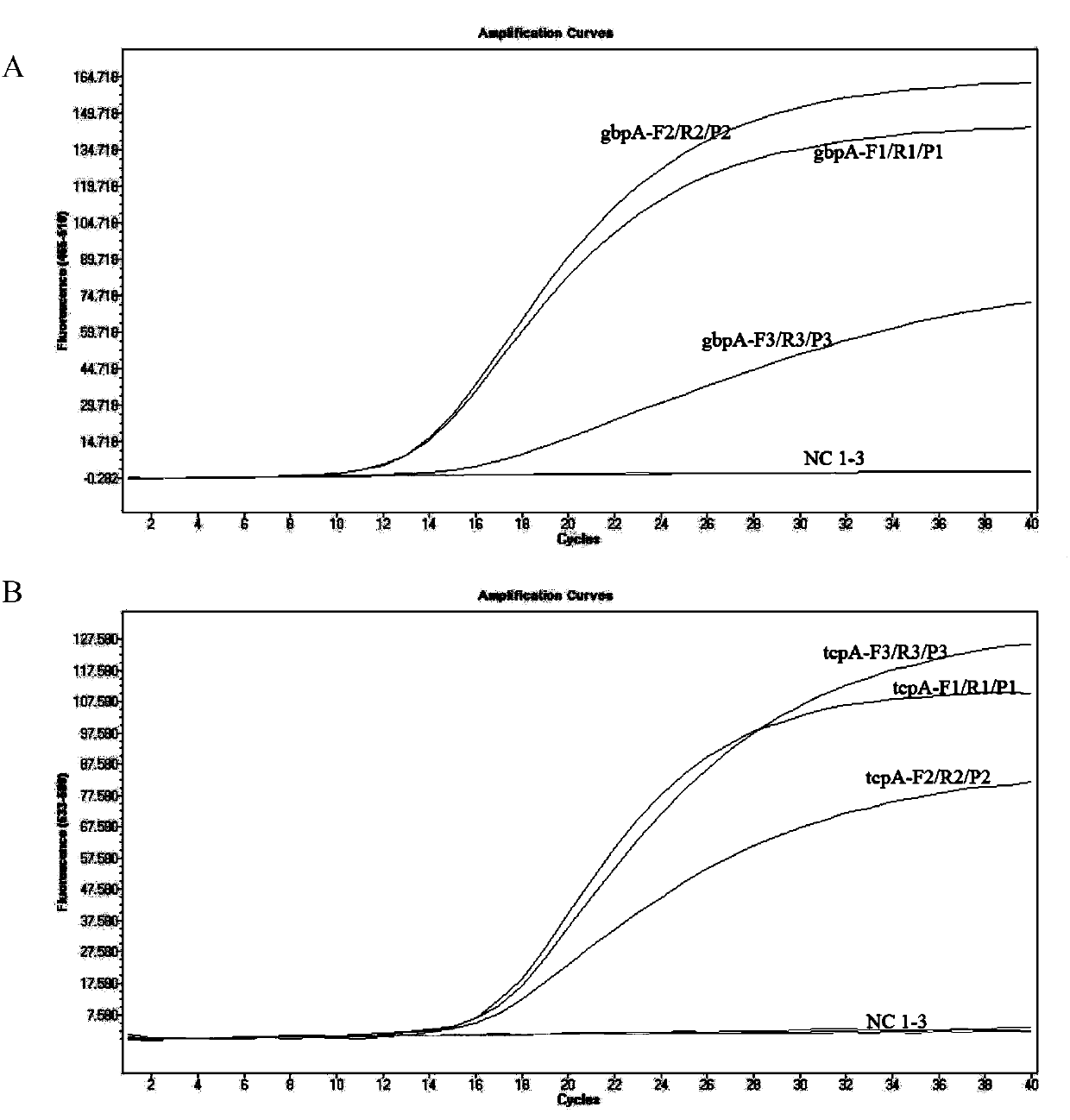
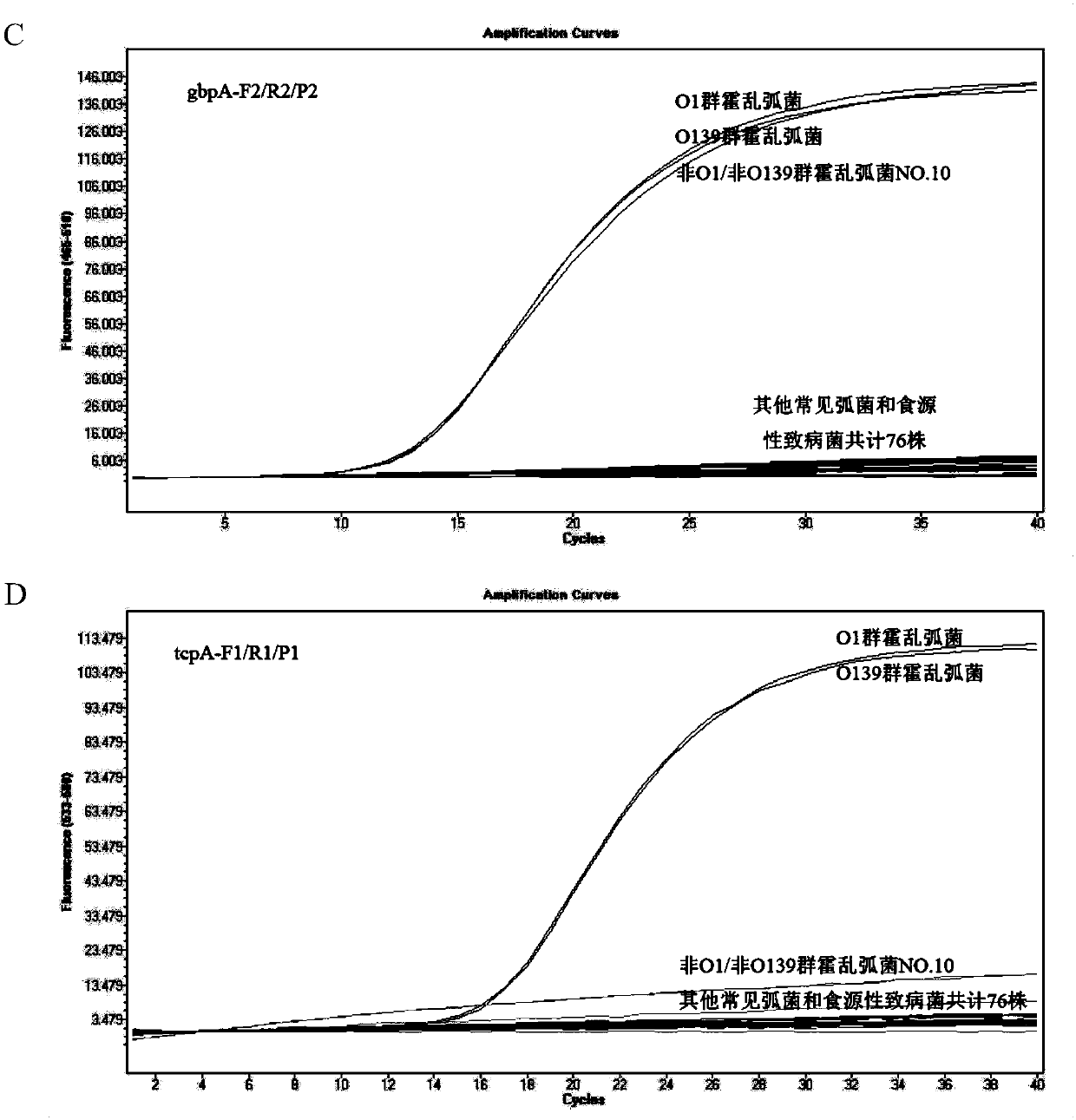
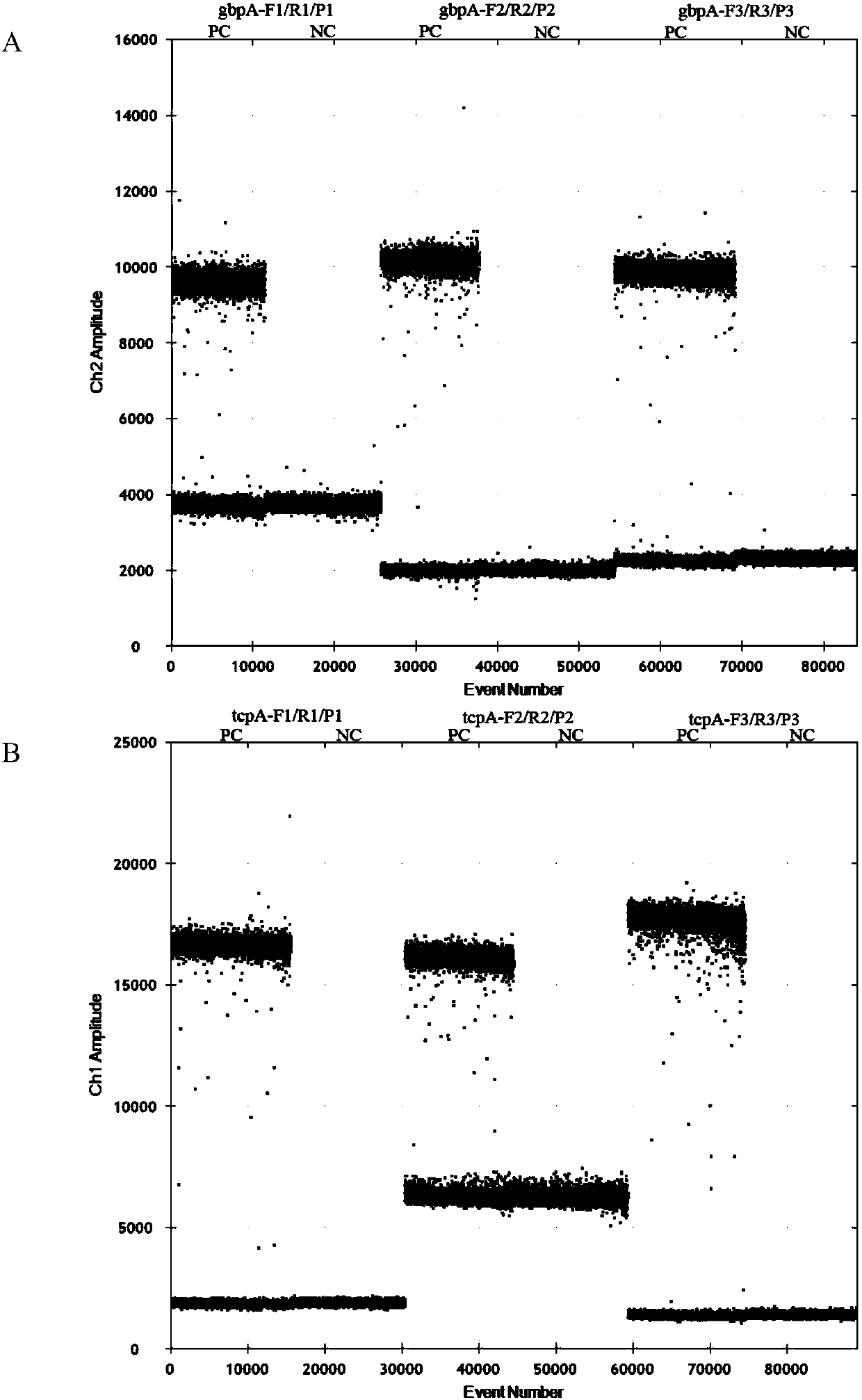
Kit and detection method for absolute quantitative detection of total vibrio cholerae and pathogenic vibrio cholerae
ActiveCN108018365AMeet the requirements of the minimum control level of pollutionReduce the impactMicrobiological testing/measurementMicroorganism based processesVibrio choleraeMicrobiology
The invention discloses a kit and a detection method for absolute quantitative detection of total vibrio cholerae and pathogenic vibrio cholerae. The present invention provides two sets of primers andprobes for the absolute quantitative detection of the total vibrio cholerae and the pathogenic vibrio cholerae. One set of the primers and probe is used for detection of the total vibrio cholera, thenucleotide sequences of the primers are as shown in SEQ ID No. 1 and SEQ ID No. 2, and the nucleotide sequence of the probe is as shown in SEQ ID No. 3. The other set of the primers and probe is usedfor detection of the pathogenic vibrio cholera, the nucleotide sequences of the primers are as shown in SEQ ID No. 4 and SEQ ID No. 5, and the nucleotide sequence of the probe is as shown in SEQ ID No. 6. The invention also provides a method for the absolute quantitative detection of the total vibrio cholerae and the pathogenic vibrio cholerae by use of a micro-droplet digital polymerase chain reaction. The method has strong specificity, high sensitivity, good amplification effect, strong operability and easy standardization.
Owner:中国海关科学技术研究中心
Micro-drop digital polymerase chain reaction chip
ActiveCN112795989AAvoid fusesAvoid breakingNucleotide librariesMicrobiological testing/measurementEngineeringBiology
The invention discloses a microdroplet type digital polymerase chain reaction chip. The chip comprises a microdroplet structure, a microdroplet collection structure and a flow channel structure; the microdroplet structure forms a sample cavity and an oil storage cavity which are spaced, the microdroplet collection structure comprises a microdroplet collection tube arranged below the microdroplet structure, a collection cavity is formed in the droplet collection pipe, the flow channel structure is arranged on the droplet structure, an outlet of the flow channel structure is communicated with the collection cavity, inlets of the flow channel structure comprise a first inlet and a second inlet, the first inlet is communicated with the lower end of the oil storage cavity, and the second inlet is communicated with the lower end of the sample cavity; an inlet of the flow channel structure is respectively communicated with the lower end of the sample cavity and the lower end of the oil storage cavity, so that a sample and an oil product can enter the flow channel structure under the action of gravity, micro-droplets wrapped with the sample are formed under the action of the flow channel structure, and an outlet of the flow channel structure is communicated with a collecting cavity, so that the micro-droplets flow into the collecting cavity through the flow channel structure; and pipetting is not needed in the whole process.
Owner:JIHUA LAB
Digital polymerase chain reaction (PCR) system
PendingCN110628610AIncrease the speed of formationReduce consumptionBioreactor/fermenter combinationsHeating or cooling apparatusThermal bubbleFormation rate
The invention provides a digital polymerase chain reaction (PCR) system. The digital PCR system comprises a droplet formation assembly and a droplet nozzle assembly; the droplet formation assembly comprises a droplet collection tank; the droplet nozzle assembly is connected under the droplet formation assembly, and comprises a plurality of droplet nozzles; the droplet nozzles communicate with thedroplet collection tank; vaporizing components are arranged inside the droplet nozzles so as to be used for vaporizing part of the liquid of a digital PCR solution in the droplet nozzles, as well as quickly pushing the remaining liquid of the digital PCR solution into droplet forming oil in the droplet collection tank so as to form digital PCR droplets. By adopting a thermal bubble technology to form high-speed digital PCR droplets, the digital PCR system is capable of achieving a droplet formation rate of more than 1000 droplets per second; and moreover, the digital PCR system has highly efficient digital PCR oil utilization rate.
Owner:SHANGHAI IND U TECH RES INST
Composition, method and system for determination of digital polymerase chain reaction (PCR)
InactiveCN110551798AImprove stabilityImprove reaction efficiencyMicrobiological testing/measurementFiberNanofiber
The invention discloses a composition, method and system for determination of a digital polymerase chain reaction (PCR). The composition comprises a water phase for generating water-in-oil liquid droplets, and the water phase contains water and fibroin nanofibers dissolved in the water. The composition particularly is an emulsion formed by the water phase and an oil phase which are mixed, whereinthe oil phase constitutes a continuous phase, and the water phase forms the multiple liquid droplets scattered in the continuous phase. The water phase contains the fibroin nanofibers, the water phasecan enter the liquid droplet interface to participate in liquid droplet formation, the liquid droplets formed by the water phase have the significantly higher reaction efficiency during the PCR, andthe stability of the liquid droplets is improved. The method for determination of the digital PCR has the high reaction efficiency and is suitable for determination of various target nucleic acids.
Owner:BEIJING ZHIYU BIOTECH LTD
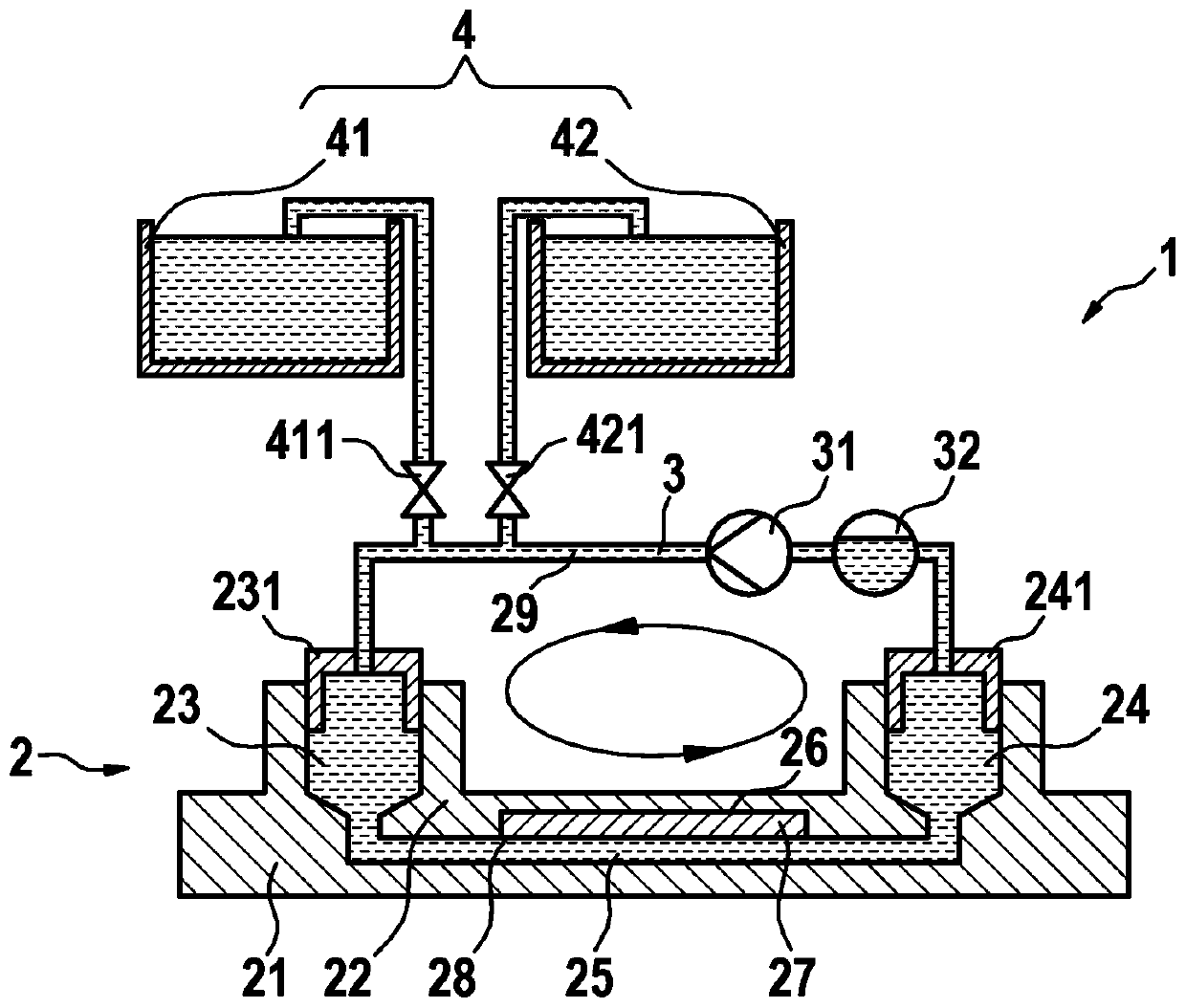
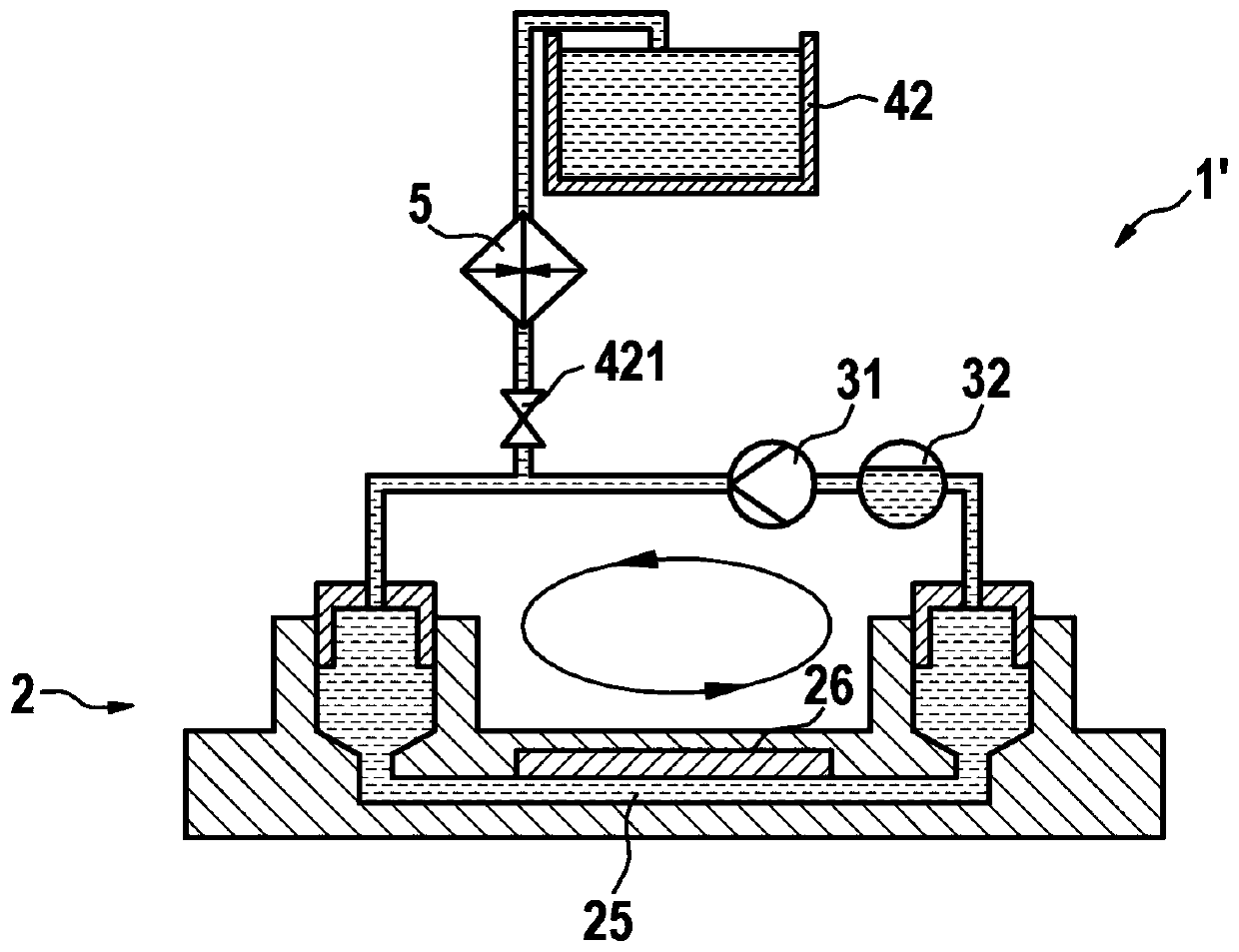
Microfluidic system for digital polymerase chain reaction of a biological sample, and respective method
PendingUS20200055042A1Heating or cooling apparatusLaboratory glasswaresPhysical chemistryEngineering
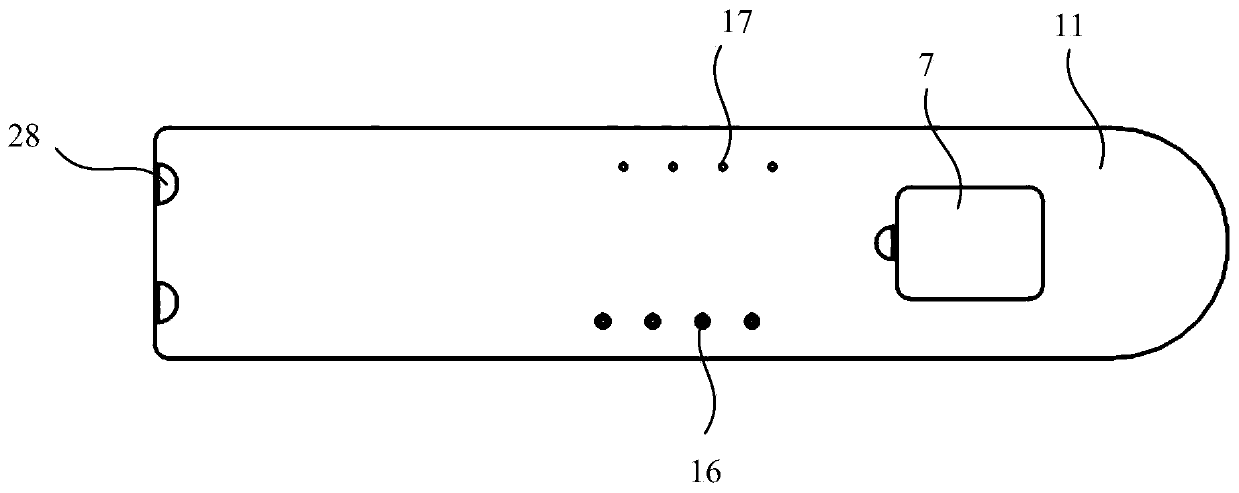
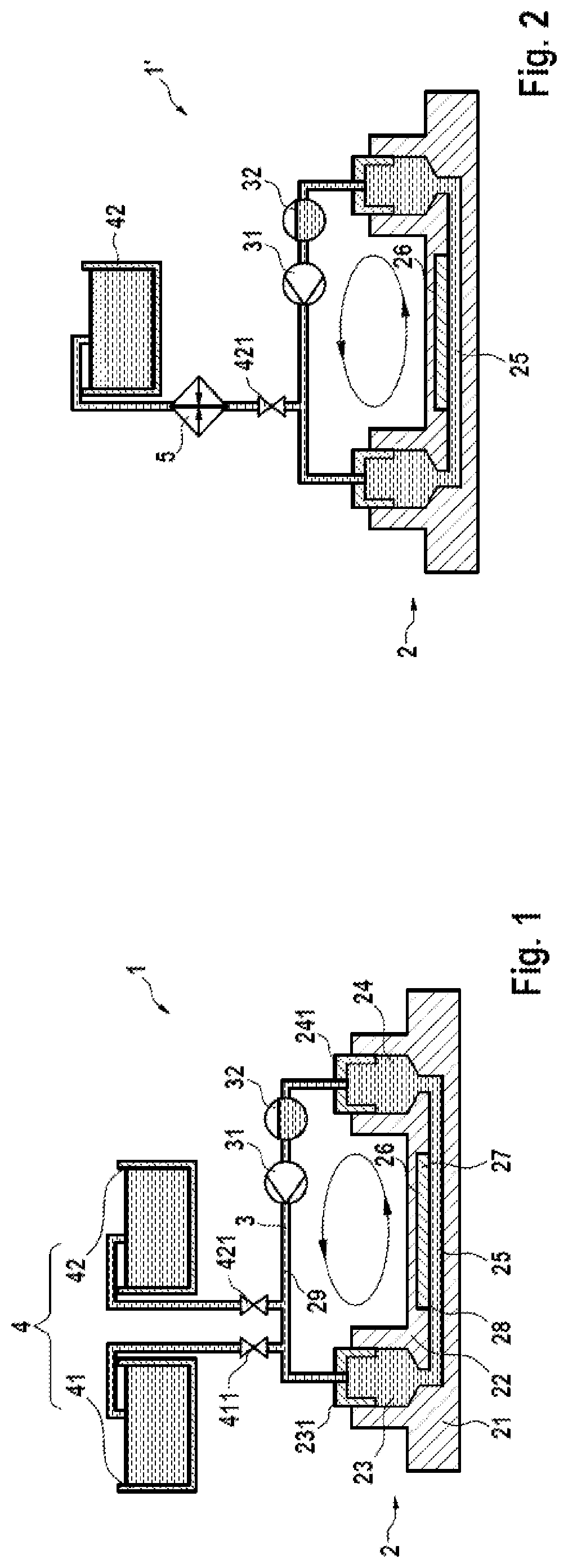
A microfluidic system (1; 1′) for dPCR of a biological sample and a respective method is provided by the present disclosure, the system (1; 1′) comprising at least one microfluidic device (2) having an inlet (23), an outlet (24), a flow channel (25) connecting the inlet (23) to the outlet (24), and an array of reaction areas (26) in fluidic communication with the flow channel (25), a flow circuit (3) connectable to the microfluidic device (2), for flowing liquid through the flow channel (25) of the microfluidic device (2), a sample liquid source connectable to the microfluidic device (2), for providing the microfluidic device (2) with a sample liquid (27), a primary sealing liquid source connectable to the microfluidic device (2), for providing the microfluidic device (2) with initial sealing liquid (28) for sealing the sample liquid (27) inside the array of reaction areas (26), a secondary sealing liquid source (4) connectable to the microfluidic device (2), for providing the microfluidic device (2) with additional sealing liquid (29), and a pumping device (31) connected to the flow circuit (3) and adapted to pump said additional sealing liquid (29) through the flow channel (25).
Owner:ROCHE MOLECULAR SYST INC
Method for detecting TCK (Tilletia controversa Kuhn) teliospores in soil through ddPCR (droplet digital polymerase chain reaction)
InactiveCN105331713AAchieving absolute quantificationSimple extraction methodMicrobiological testing/measurementTilletia ayresiiSpins
The invention relates to a method for detecting TCK (Tilletia controversa Kuhn) teliospores in soil through ddPCR (droplet digital polymerase chain reaction). The method comprises steps as follows: (1), total DNA (deoxyribonucleic acid) of a to-be-detected soil sample is extracted; (2), the total DNA of the to-be-detected soil sample serves as a template for ddPCR, and a PCR product is obtained; (3), the PCR product is put in a droplet read instrument, a signal is read, experimental data are analyzed, and the absolute content of TCK in the soil sample is obtained. A total DNA extraction method comprises steps as follows: the taken-back soil sample is sieved by a mesh screen for impurity removal and then is frozen; total DNA of the soil sample is extracted by adopting FastDNA<TM> SPIN Kit for Soil. With the adoption of the method, higher-quality DNA can be extracted from the soil, ddPCR is adopted for detection, accordingly, the absolute quantification of TCK in the soil is realized, control measures are taken in time, and economic losses are reduced.
Owner:INST OF PLANT PROTECTION CHINESE ACAD OF AGRI SCI
Methods and systems for volume variation modeling in digital PCR
A method for performing digital polymerase chain reaction (dPCR) is provided. The method includes partitioning a biological sample volume including a plurality of target nucleic acids into a plurality of partitions, where at least one partition includes at least one target nucleic acid. The method further includes determining a model for volume variation of the plurality of partitions and determining a number of partitions including at least one target nucleic acid. The method includes generating a concentration of target nucleic acids in the biological sample based on the model for volume variation and the fraction of partitions including at least one target nucleic acid.
Owner:LIFE TECH CORP
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com