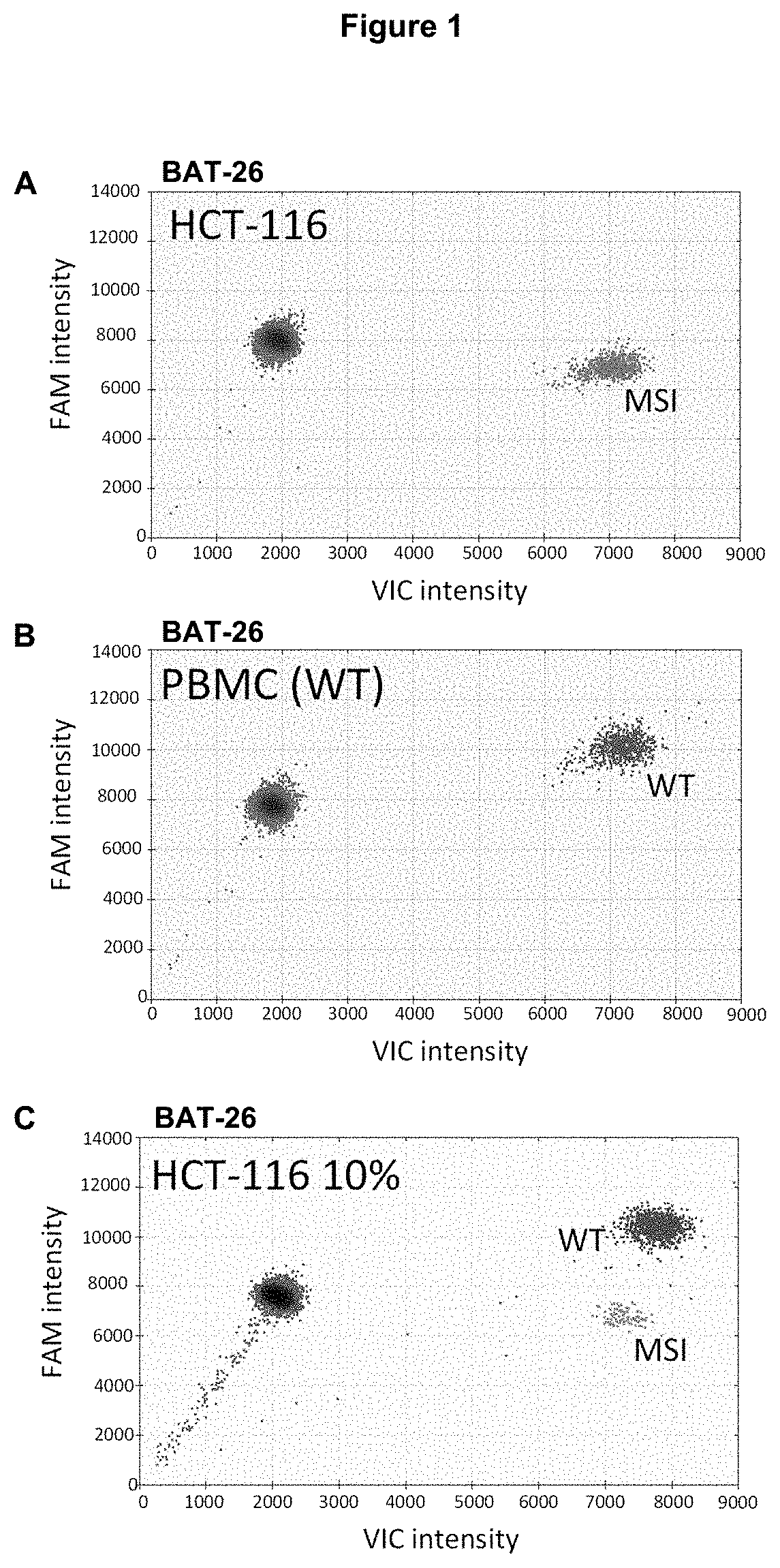
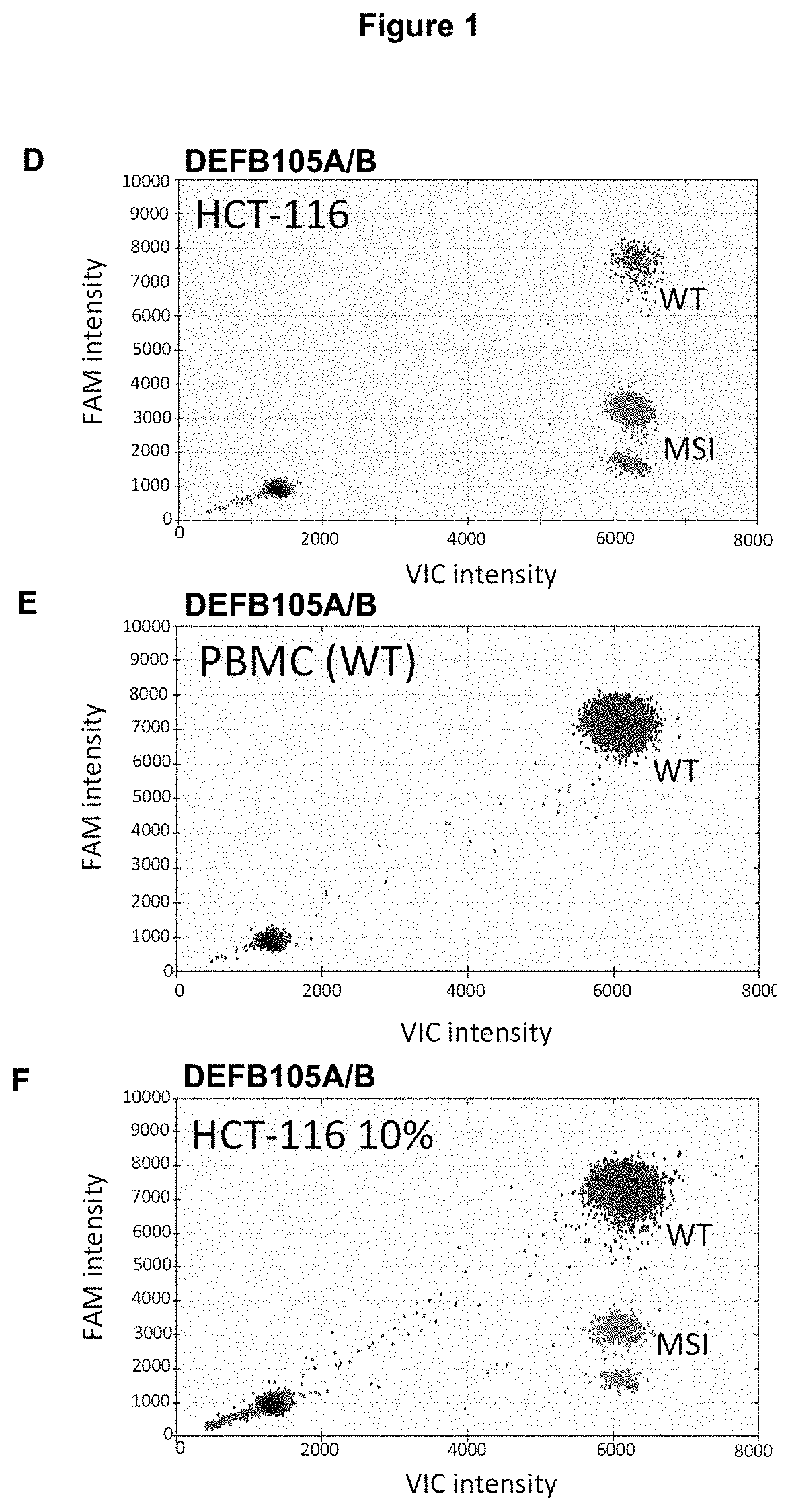
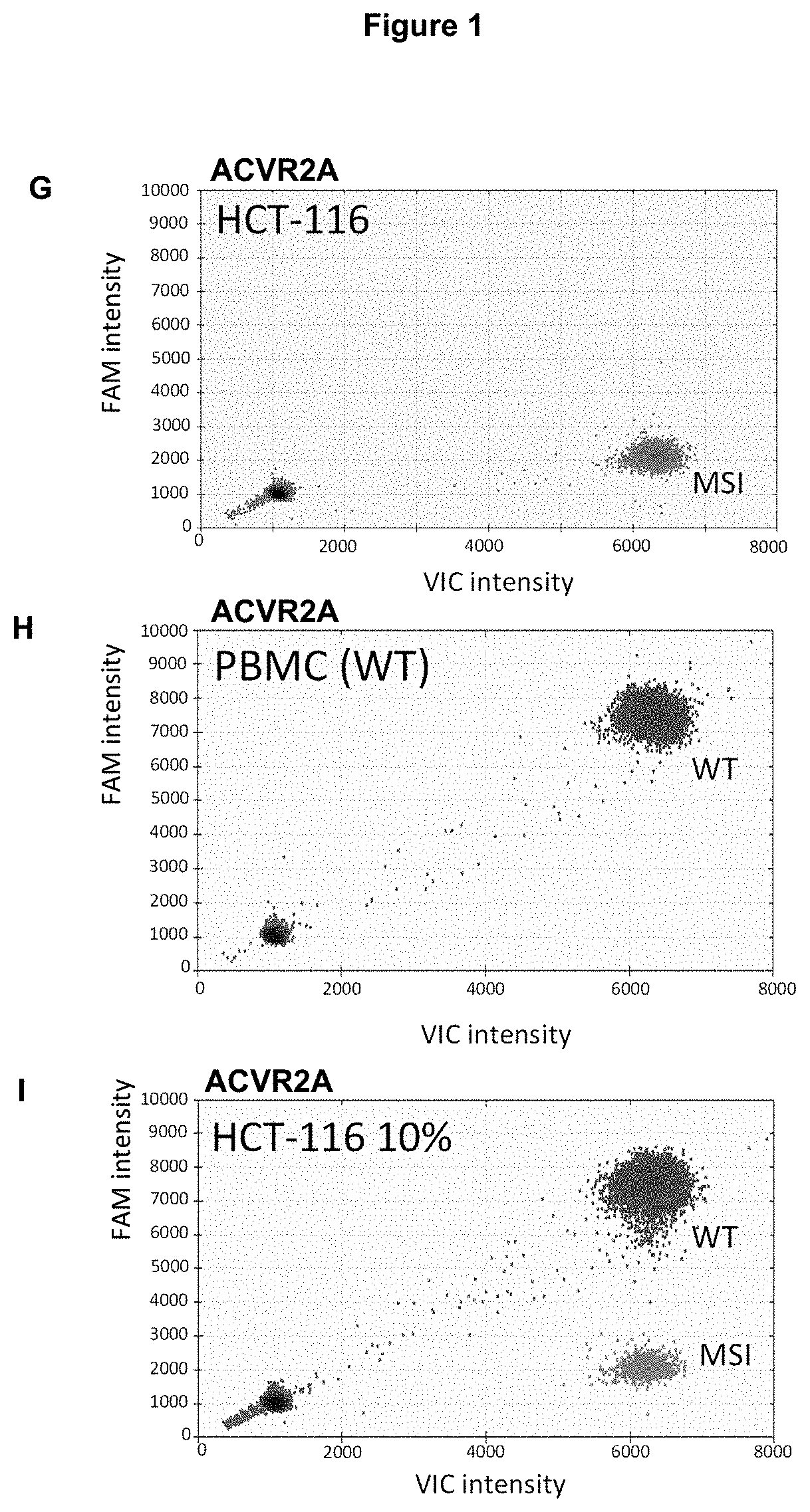
Method for Detecting a Mutation in a Microsatellite Sequence
a microsatellite sequence and mutation technology, applied in the field of microsatellite sequence mutation detection, can solve the problems of inability to efficiently or reliably hybridize the probe, the use of such a technique has never been envisioned for the detection of a mutated microsatellite sequence, and the inability to discriminate between wt and mutant microsatellite sequences, etc., to achieve the effect of increasing sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0035]A—Definitions
[0036]The following definitions are intended to assist in providing a clear and consistent understanding of the scope and detail of the following terms, as used to describe and define the present invention:
[0037]As used herein, the verb “comprise” as is used in this description and in the claims and its conjugations are used in its non-limiting sense to mean that items following the word are included, but items not specifically mentioned are not excluded. In addition, reference to an element by the indefinite article “a” or “an” does not exclude the possibility that more than one of the elements are present, unless the context clearly requires that there is one and only one of the elements. The indefinite article “a” or “an” thus usually means “at least one.”
[0038]As used herein a “tumor” or a “neoplasm” (both terms can be used interchangeably) is an abnormal new growth of cells. The cells in a neoplasm usually grow more rapidly than normal cells and will continue...
PUM
| Property | Measurement | Unit |
|---|---|---|
| fluorescence | aaaaa | aaaaa |
| fluorescence intensity | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com