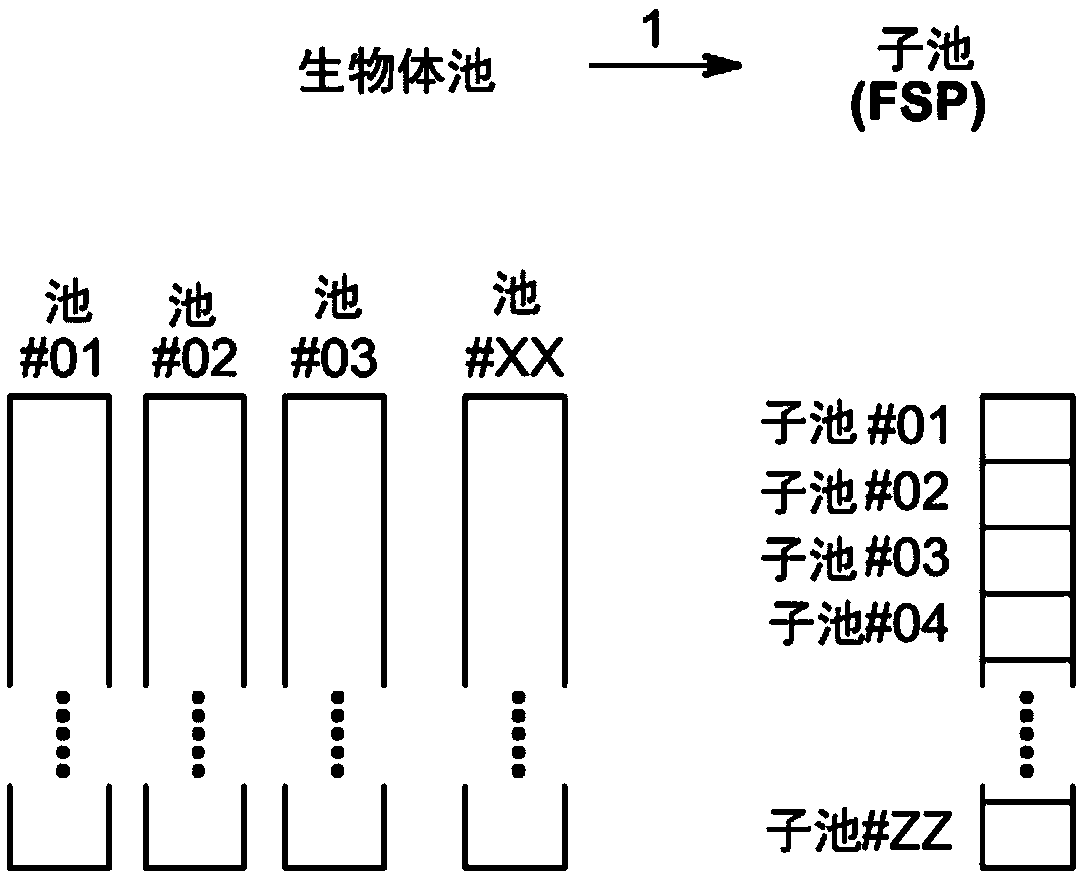
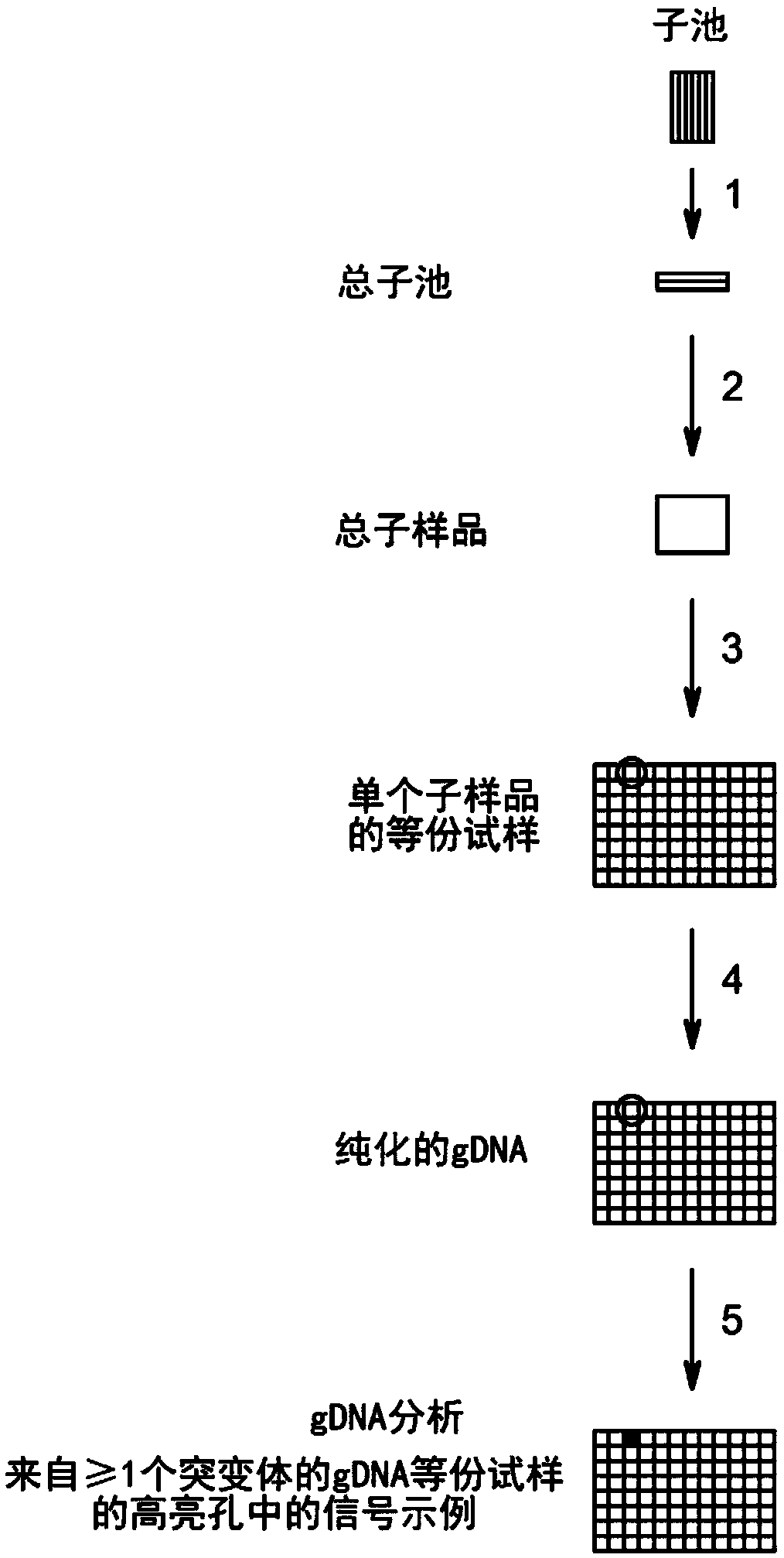
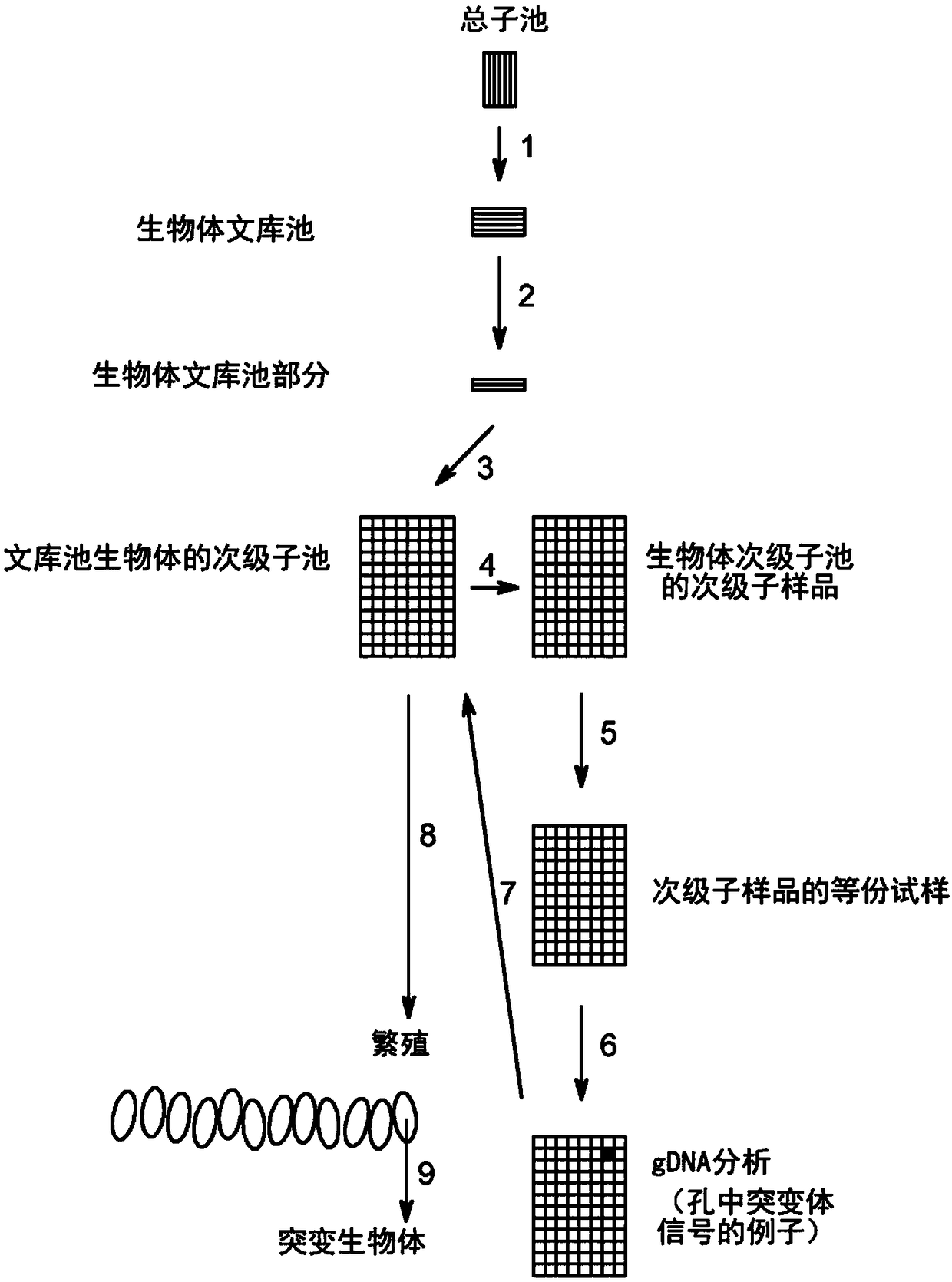
Method to screen for a mutant within a population of organisms by applying a pooling and splitting approach
A technology of organisms and species, applied in the direction of mutant preparation, microbial measurement/inspection, microbial library, etc., can solve unrealistic problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0484] - Example 1: Growing barley plants in "fields";
Embodiment 2
[0485] - Example 2: Harvesting of grain in a single "plot".
[0486] WS3: Determine if a library sample contains mutant grain
[0487] Step 3.1: Divide a single "Total Grain" grain pool into two parts
[0488] The threshed grain in a single "Total Grain" bag was divided into two parts, where the "Total Sub-Grain" sample consisted of ~1500 kernels (labeled SGT#01, SGT#02, etc.; see Figure 2C ; Action 1), its processing details refer to the following steps 3.2 to 3.4. Broadly speaking, grain processing in the "Total Grain Grain" section seeks to determine which fraction of the grain contains the mutation of interest. For reasons of clarity, but irrelevant to the associated actions of step 3, the remainder of the "total amount of grains" consists of ~4500 kernels, which are processed as detailed in the description relating to step 4.
[0489] For purposes of clarity of description, and not by way of limitation, the following Step 3.1 related subject matter is detailed in the ...
Embodiment 3
[0490] - Example 3: Description of the grains in "Total amount of sub-grains".
[0491] Step 3.2: Preparation of flour samples from the grains of the "Total Grain Grains"
[0492] Total samples of each sub-grain containing 1500 grains, ie samples designated as SGT#01, SGT#02, etc., were ground in a standard laboratory grinder (see Figure 2B , action 2), with thorough cleaning between applications, resulting in the corresponding flour samples - denoted as SFT#01, SFT#02, etc. (see Figure 2C ).
[0493] For purposes of clarity of description, and not by way of limitation, Step 3.2-related subject matter below is detailed in the following examples:
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com