Kit and detection method for absolute quantitative detection of total vibrio cholerae and pathogenic vibrio cholerae
An absolute quantification technique for Vibrio cholerae, applied in the field of molecular biology, can solve the problems that the validity and practicability of the quantitative analysis of Vibrio cholerae are unknown, the quantitative detection of Vibrio cholerae has not been seen, and the composition of the background bacteria is complex, etc., to achieve The effect of shortening detection time, avoiding deviation, and reducing influence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2d
[0046] The design of embodiment 2ddPCR primers, probes
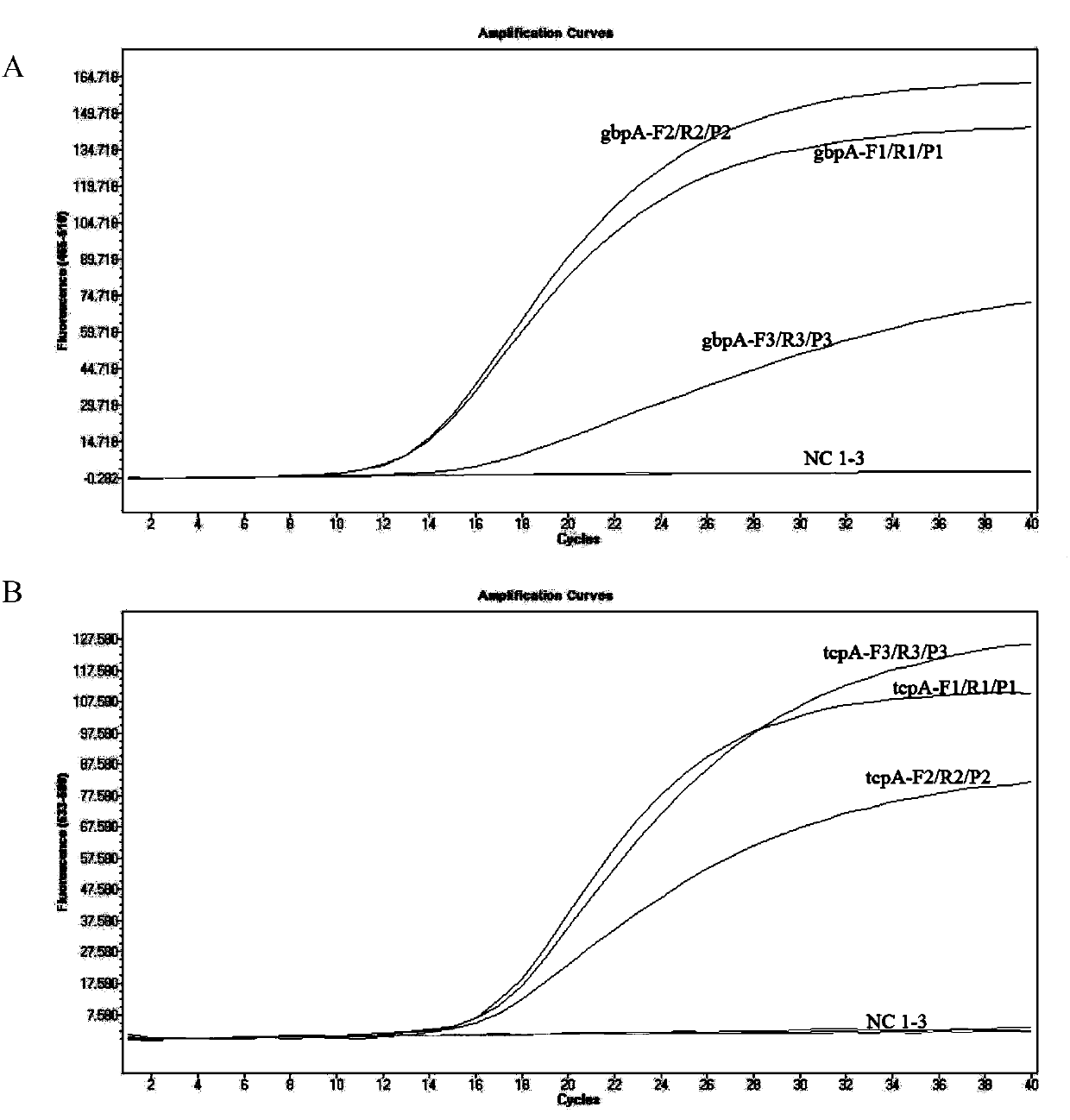
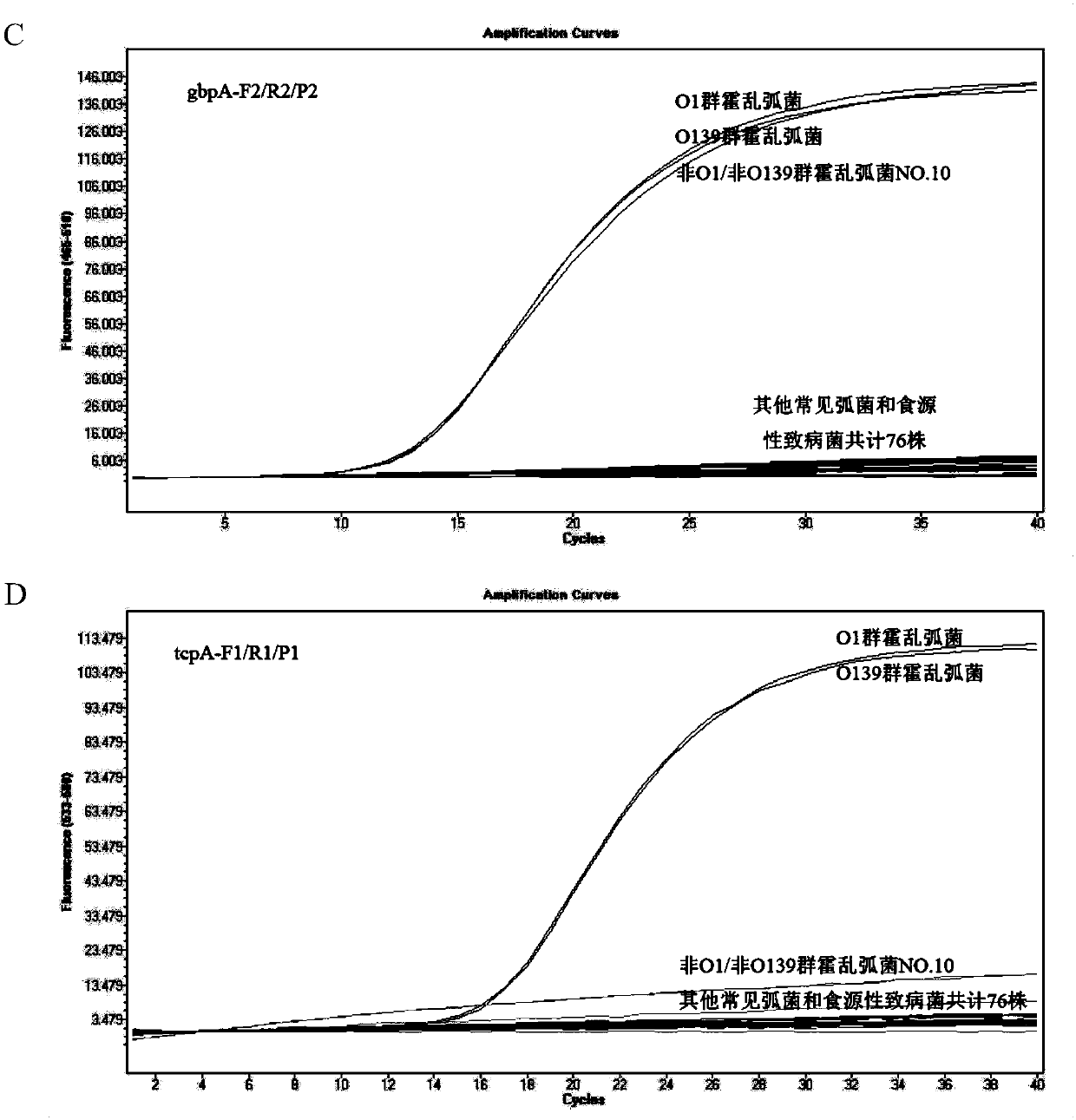
[0047] In order to achieve the specific detection and absolute quantitative analysis of total Vibrio cholerae and pathogenic Vibrio cholerae, we selected a single copy of species-specific and highly conserved N-acetylglucosamine (GlcNAc) binding protein A The coding gene gbpA (GenBank no.EU072441.1) and the virulence co-regulatory pilus coding gene tcpA (GenBank no.KP187623.1) were used as target sequences, and the sequences were analyzed and compared through NCBI online tools, using Prime Express software V3.0 (ABI, Foster City, CA, USA) designed three pairs of primers and probes, and the sequences are shown in Table 1.
Embodiment 3d
[0048] Example 3 ddPCR detection
[0049] 1. Preparation of reaction premix
[0050] Prepare 20 μL ddPCR reaction master mix according to Table 2.
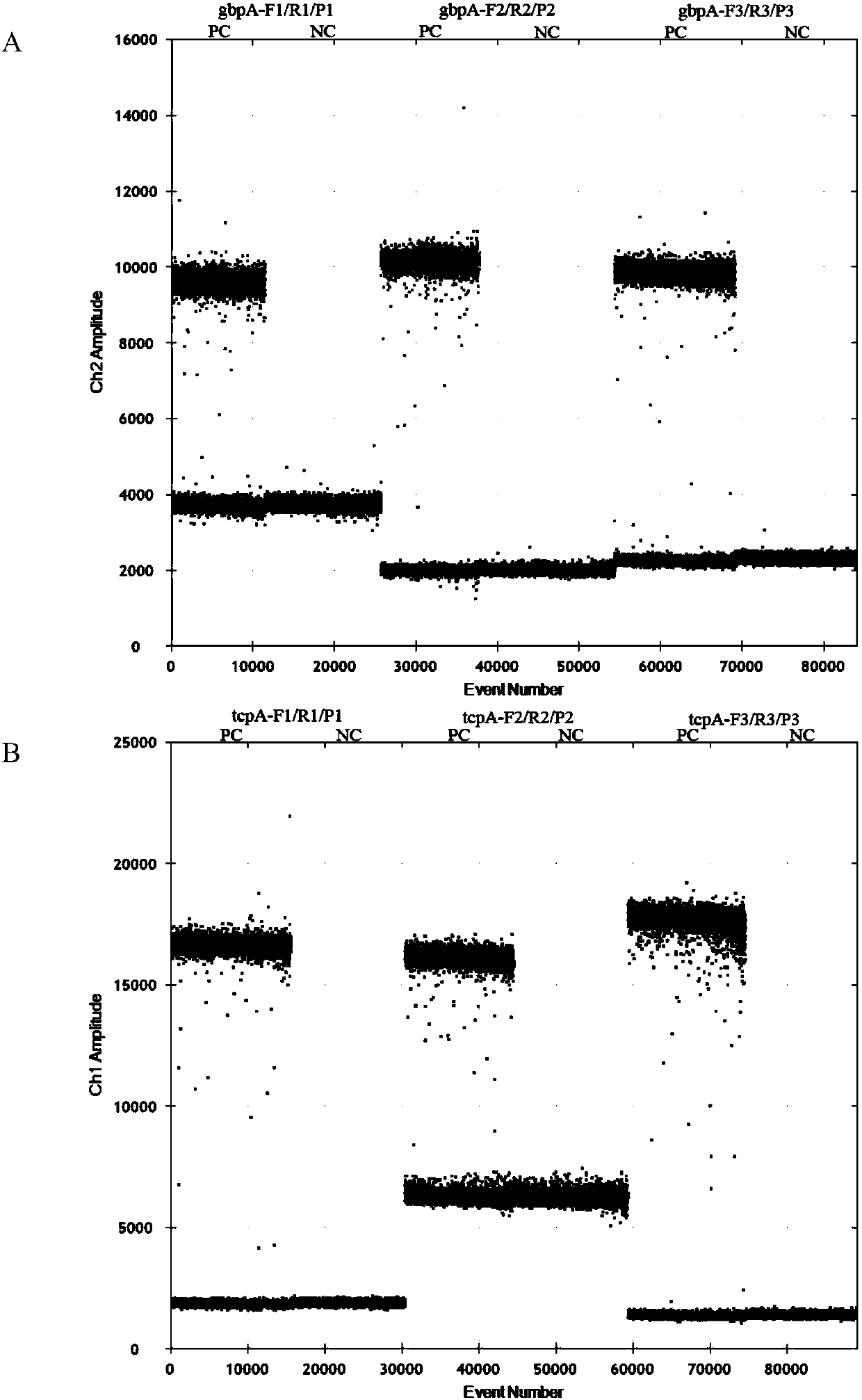
[0051] 2. Droplet generation
[0052] 20 μL of ddPCR reaction master mix and 70 μL of droplet generating oil were added to the 8-well droplet generating card, and placed in a droplet generator (QX100, Bio-Rad, Pleasanton, CA) to generate droplets.
[0053] 3. Amplification reaction
[0054] The generated water-in-oil droplets (40 μL) were slowly transferred to a 96-well plate, sealed and placed on a PCR machine (GeneAmp9700, Applied BioSystems, Foster City, CA) for amplification reaction. The amplification conditions are shown in Table 3 shown.
[0055] 4. Result judgment
[0056] The amplified 96-well plate was placed in a droplet reader (QX100, Bio-Rad, Pleasanton, CA), and the results were read and analyzed using QuantaSoft software. Positive microdroplets containing amplified products and negative microdroplets without a...
Embodiment 4
[0057] Embodiment 4qPCR detects
[0058] In order to facilitate comparison with the ddPCR method, the same set of primers and probes were selected for qPCR detection. The 25 μL double qPCR reaction system includes: 2×Master Mix 12.5 μL, 10 μM gbpA-F, gbpA-R, tcpA-F and tcpA-R 1 μL each, 10 μM gbpA-P and tcpA-P 0.5 μL each, to be tested DNA template 2 μL. The amplification was carried out on ABI7900 fluorescent PCR instrument (US, ABI company) or Light cycler 480II (Switzerland, Roche company), 50°C for 2min, 95°C for 10min, 95°C for 15s, 60°C for 1min, 45 cycles. After the reaction is completed, use SDS3.2 or SW1.5.1 software for analysis, and calculate the copy number of the target gene according to the standard curve.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com