Kit and detection method for absolute quantitative detection of vibrio parahaemolyticus
A hemolytic Vibrio, absolute quantitative technology, applied in the field of molecular biology, can solve the problems of time-consuming, high concentration, complex background bacteria, etc., achieve high accuracy and repeatability, simplify the operation process, and shorten the detection time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0047] Example 2 ddPCR primer, probe design, screening and specificity verification
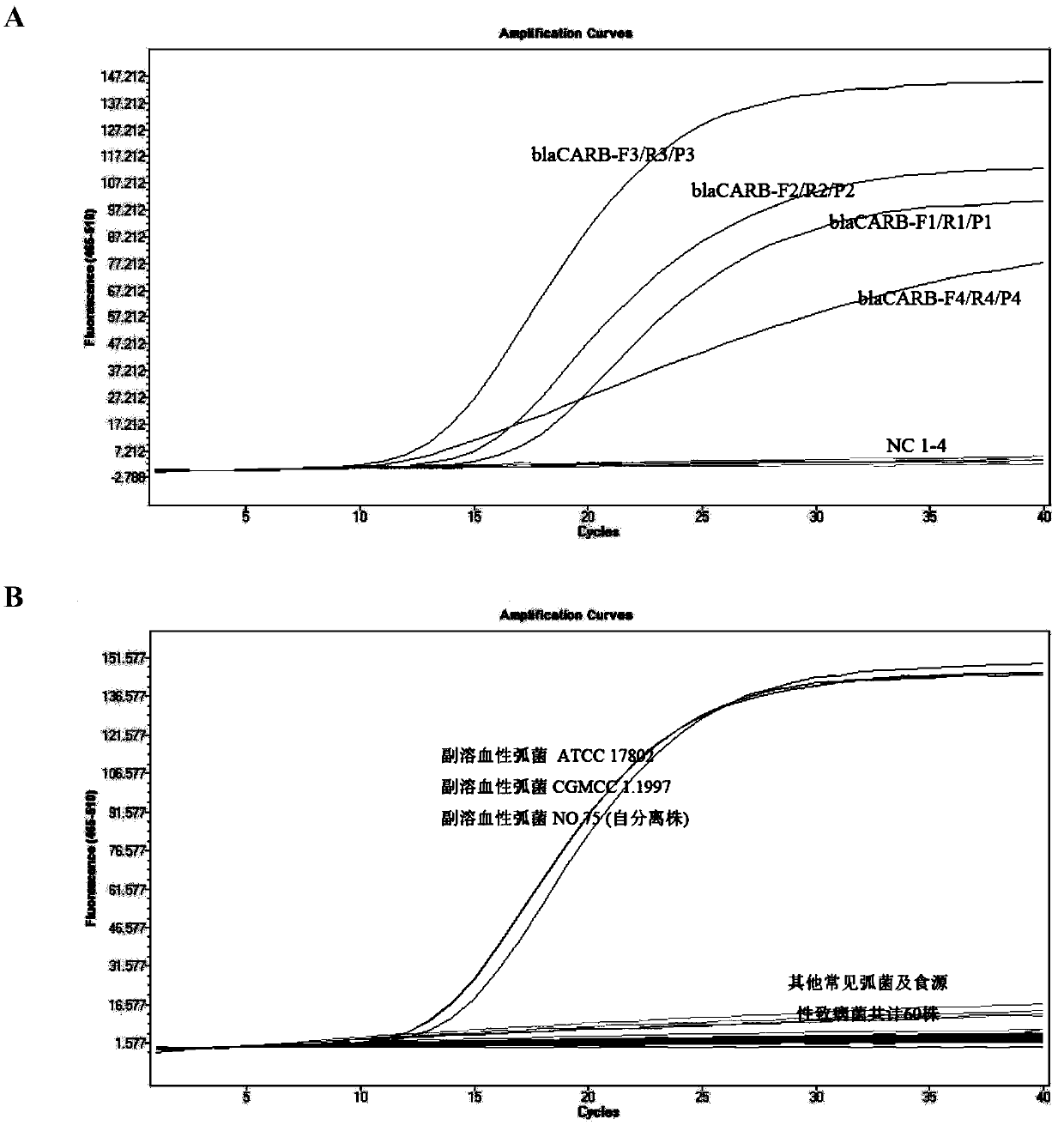
[0048] In order to achieve the specific detection and absolute quantitative analysis of Vibrio parahaemolyticus, we selected a species-specific and highly conserved single-copy β-lactamase coding gene blaCARB (GenBank no.NG_048735.1 ) as the target sequence, sequence analysis and alignment were performed by NCBI online tools, and 4 pairs of primer and probe combinations were designed using Prime Express software V3.0 (ABI, Foster City, CA, USA). The sequences are shown in Table 1. The 5' end of the probe is labeled with FAM, and the 3' end is labeled with BHQ. Primers / probes were synthesized by Beijing Liuhetong Economic and Trade Co., Ltd.
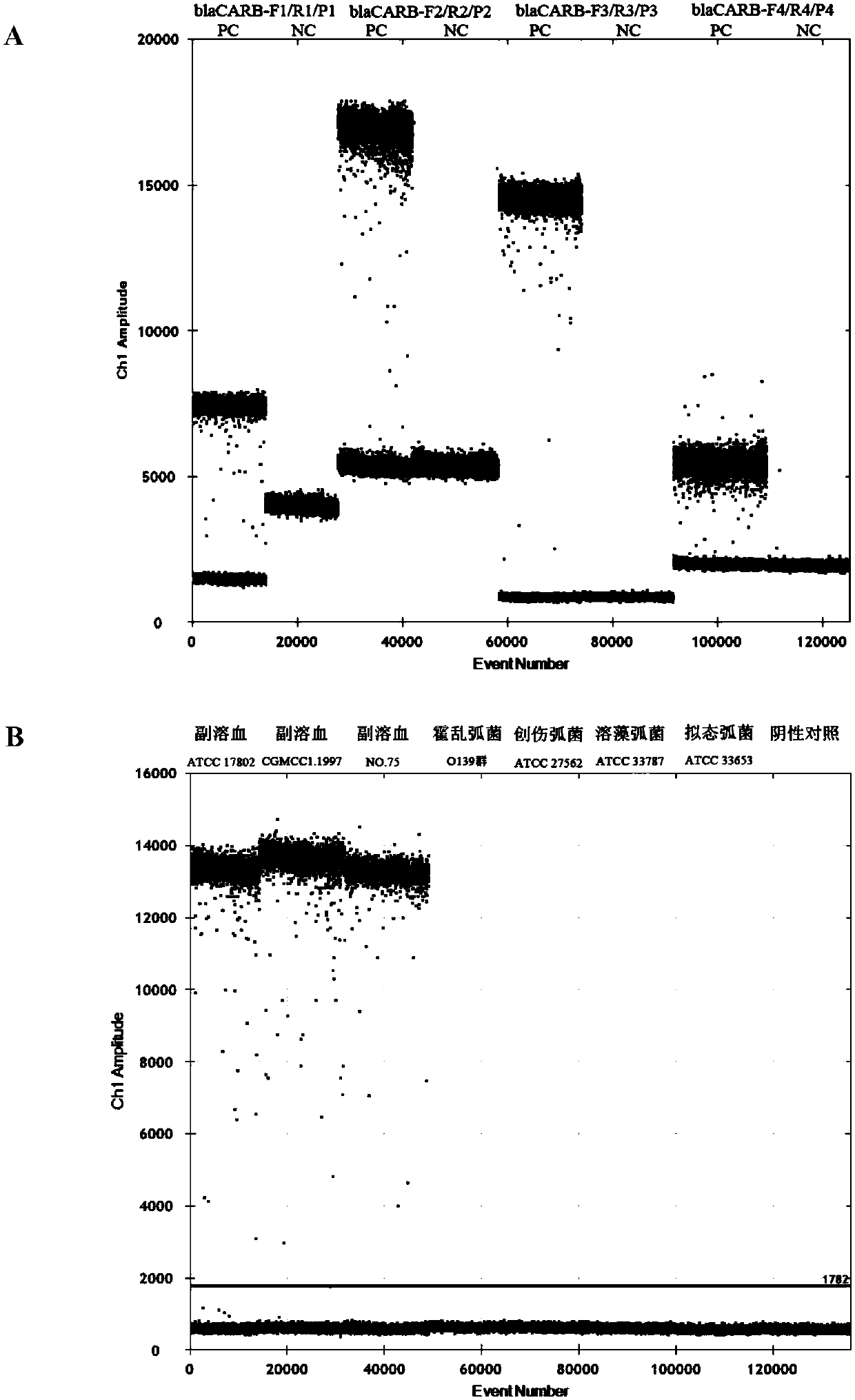
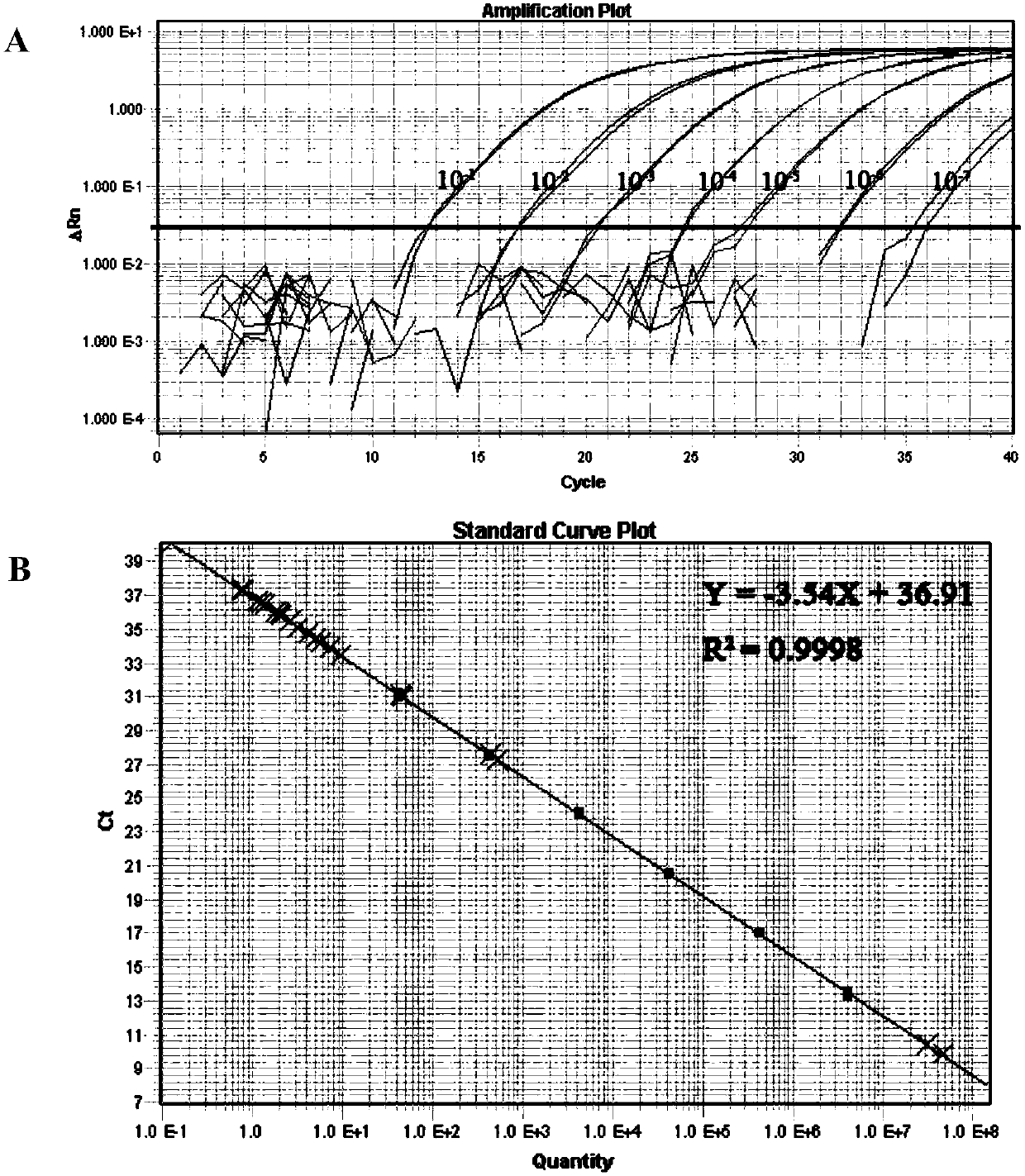
[0049] First, the qPCR method was used to screen the 4 pairs of primers and probes that had been designed and synthesized. Through the detection of the genomic DNA of Vibrio parahaemolyticus (ATCC17802), it was found that all the primer probes could ef...
Embodiment 3
[0051] Embodiment 3 detects the ddPCR kit of vibrio parahaemolyticus
[0052] The kit mainly includes:
[0053] (1) Primers and probes for detecting Vibrio parahaemolyticus (the sequences of the primers and probes are shown in SEQ ID No.1-3), synthesized by Beijing Liuhetong Economic and Trade Co., Ltd.;
[0054] (2) Positive control: Genomic DNA of Vibrio parahaemolyticus ATCC 17802, 1000 copies / μL;
[0055] (3) ddPCR master mix, purchased from BioRad, USA;
[0056] (4) microdroplet generating oil, purchased from BioRad Company of the United States;
[0057] (5)) Droplet generation card, purchased from BioRad, USA;
[0058] (6) Twin Tec Semi-Skirted 96-well plate, purchased from Eppendorf, Germany;
[0059] (7) Aluminum foil heat-sealing film, purchased from American BioRad Company.
Embodiment 4
[0060] The establishment of embodiment 4 detection method
[0061] (1) Extract the DNA of the genome of the sample to be tested: use the Bacterial Genome DNA Extraction Kit (Tiangen Biochemical Technology Co., Ltd., article number: DP302) to extract the DNA of each dilution of the bacterial liquid, and the extraction steps are carried out according to the kit instructions, and finally the nucleic acid is dissolved In 50 μL TE buffer;
[0062] (2) Configure the ddPCR reaction system, see Table 2 for details;
[0063] (3) Droplet generation: Add 20 μL of ddPCR reaction master mix and 70 μL of droplet generation oil to the 8-well droplet generation card, and place it in a droplet generator (QX100, Bio-Rad, Pleasanton, CA) to generate droplet;
[0064] (4) Amplification reaction: Slowly transfer the generated water-in-oil droplets (40 μL) to a 96-well plate, seal the film and place on a PCR machine (GeneAmp 9700, Applied BioSystems, Foster City, CA) for amplification Reaction, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com