Nucleic acid combination for detecting pseudorabies virus and application of nucleic acid combination, and kit and method for detecting pseudorabies virus
A technology of pseudorabies virus and kit, applied in the direction of microorganism-based methods, biochemical equipment and methods, recombinant DNA technology, etc., can solve the problems of difficult and accurate collection of trigeminal ganglion, missed detection, etc., achieve high sensitivity, avoid The effect of missing detection and improving the detection rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
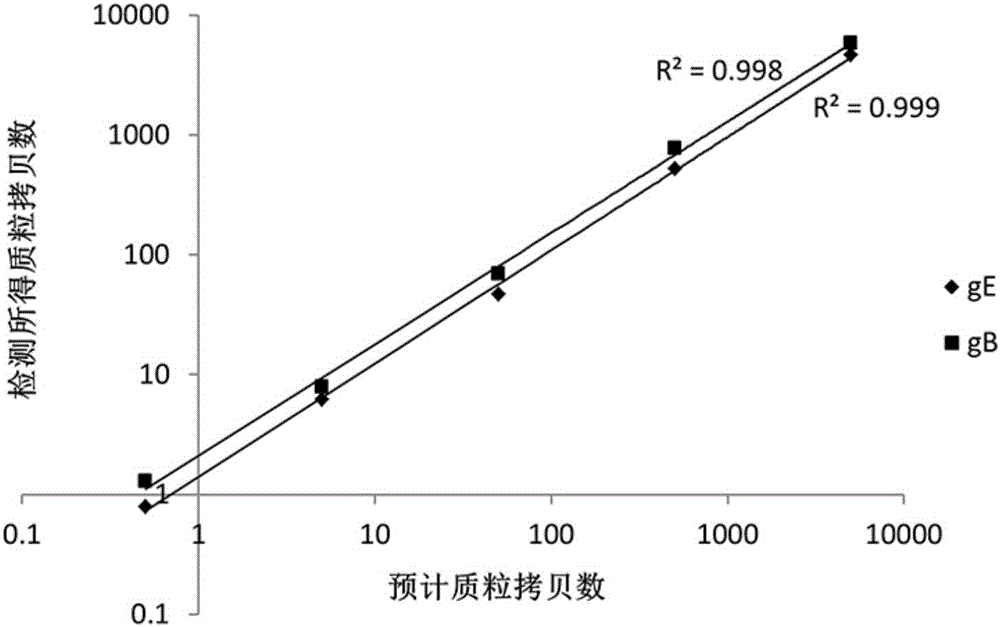
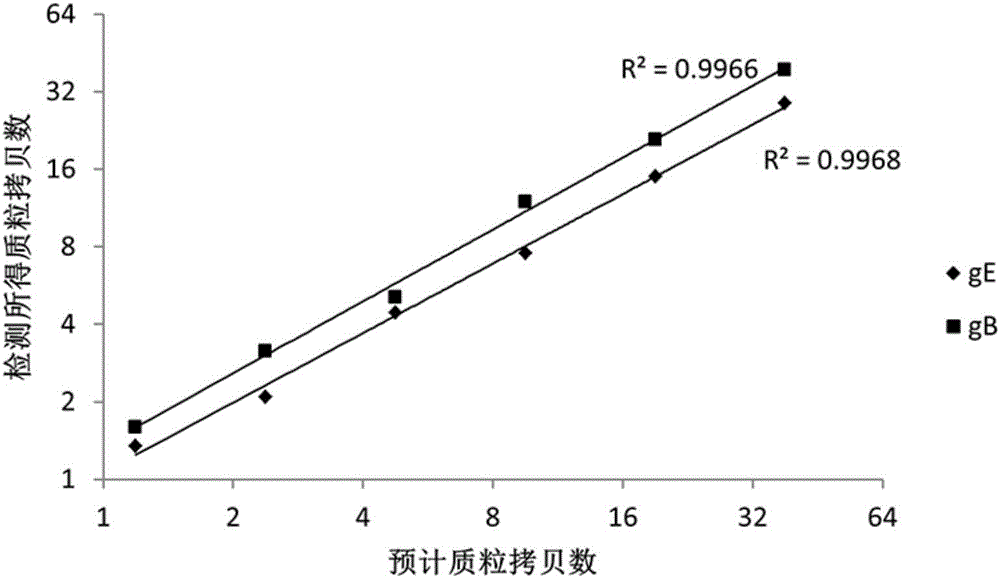
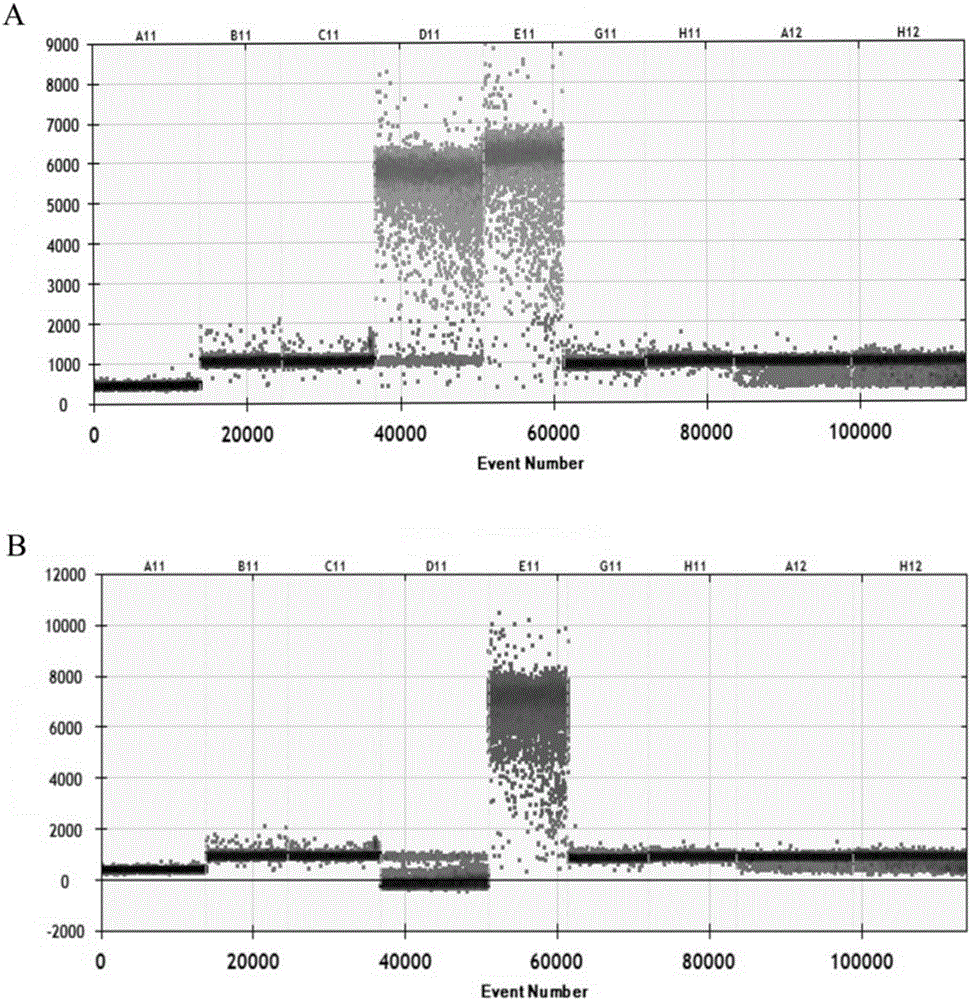
[0052] The present embodiment provides a nucleic acid combination for detecting pseudorabies virus, which includes a first primer pair and a first probe corresponding to the first primer pair, and a second primer pair and a second probe corresponding to the second primer pair. Needle. The first pair of primers and the first probe are designed according to the conserved sequence of the gB gene of pseudorabies virus, and can be used to detect whether the sample contains pseudorabies virus. The second primer pair and the second probe are designed according to the conserved sequence of the gE gene of pseudorabies virus. By adding the second primer pair and the second probe, it is possible to further distinguish all pseudorabies viruses while detecting whether the sample contains pseudorabies virus. Whether the detected pseudorabies virus is a field strain (gB positive, gE positive) or a gE-deleted commercial vaccine strain (gB positive, gE negative).
[0053] It should be noted t...
Embodiment 2
[0061] This embodiment provides a kit for detecting pseudorabies virus, which includes the nucleic acid combination provided in Embodiment 1. The kit can be used to detect whether a sample contains pseudorabies virus on the ddPCR platform, and at the same time distinguish the detected pathogen, that is, whether the detected pseudorabies virus is a wild strain or a vaccine strain with gE gene deletion, Moreover, the kit provided in this embodiment has good sensitivity and specificity, and can avoid the occurrence of missed detection caused by the small amount of virus contained in the sample.
[0062] Of course, in other embodiments, the kit for detecting pseudorabies virus provided by the present invention may also include one or more of PCR reaction buffer, dNTP, Taq DNA polymerase and the like when performing ddPCR.
Embodiment 3
[0064] This embodiment provides a method for detecting pseudorabies virus, specifically as follows.
[0065] 1. Prepare ddPCR reaction system: 10 μL of 2×ddPCR master mix, 0.9 μL of each primer (900nM) (0.9 μL each of gB-F, gB-R, gE-F, gE-R), and 0.5 μL of each probe (250nM) (0.5 μL each for gB-probe and gE-probe), 2 μL of the DNA template of the sample to be tested, and make up to 20 μL with DEPC-treated water.
[0066] The base sequences of gB-F, gB-R, gE-F, gE-R, gB-probe, and gE-probe are the same as in Example 1.
[0067] 2. Generate microdroplets: microdroplets are formed by wrapping ddPCR reaction system with microdroplet generating oil, the volume ratio of microdroplet generating oil to ddPCR reaction system is 7:2, and the number of microdroplets is 1×10 4 indivual. In this example, the ddPCR reaction system was coated with 75 μL of droplet generating oil on a ddPCR instrument (QX100 microdroplet digital PCR instrument, Bio-Rad, USA) to form microdroplets.
[0068]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com