Method for detecting ARMS-ddPCR (amplification refractory mutation system-droplet digital polymerase chain reaction) gene site-directed mutation
A detection method and point mutation technology, applied in the field of gene point mutation detection methods, can solve the problems of long optimization time, small dynamic range, false positives, etc., and achieve the effects of high sensitivity, simple optimization and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
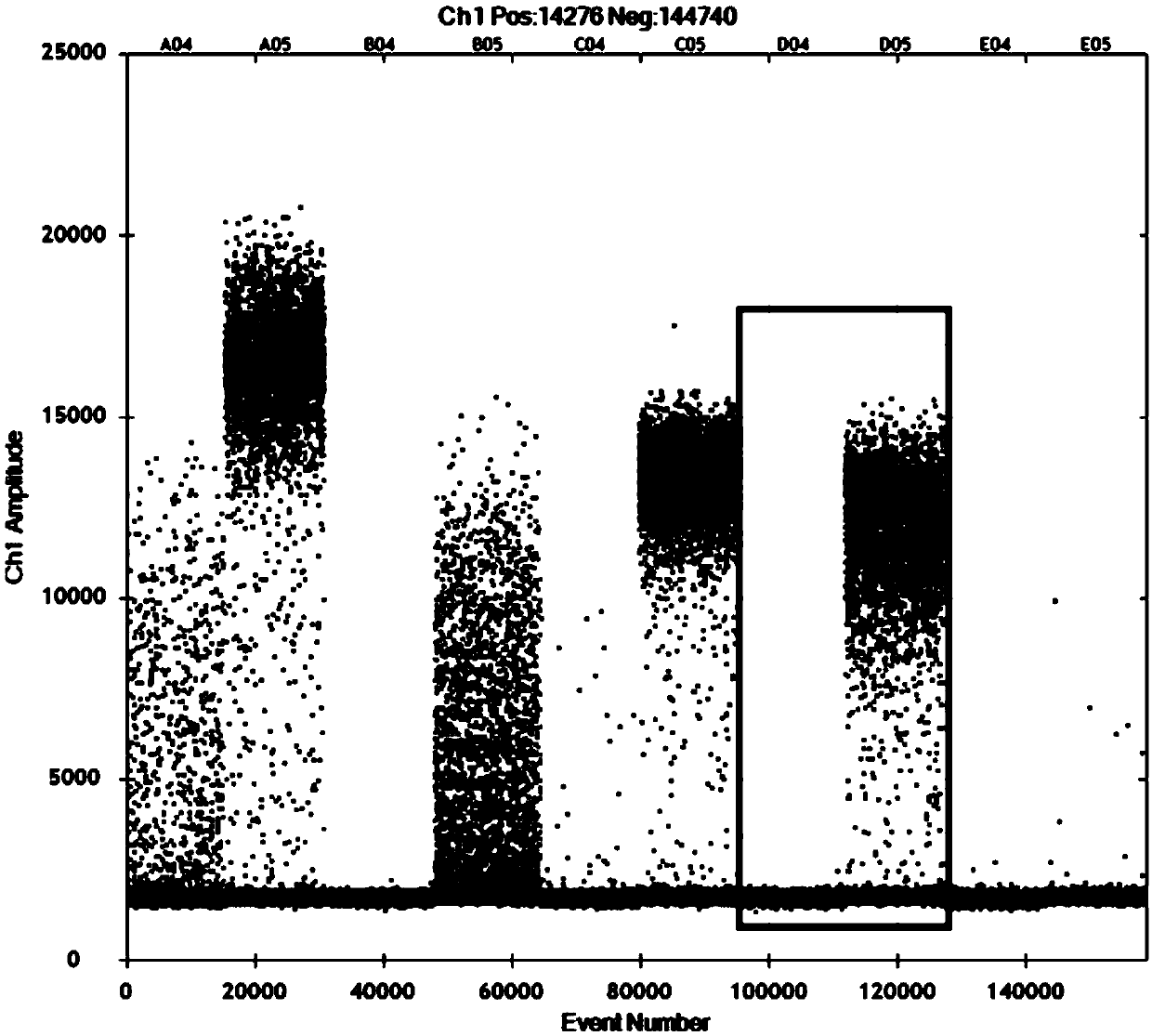
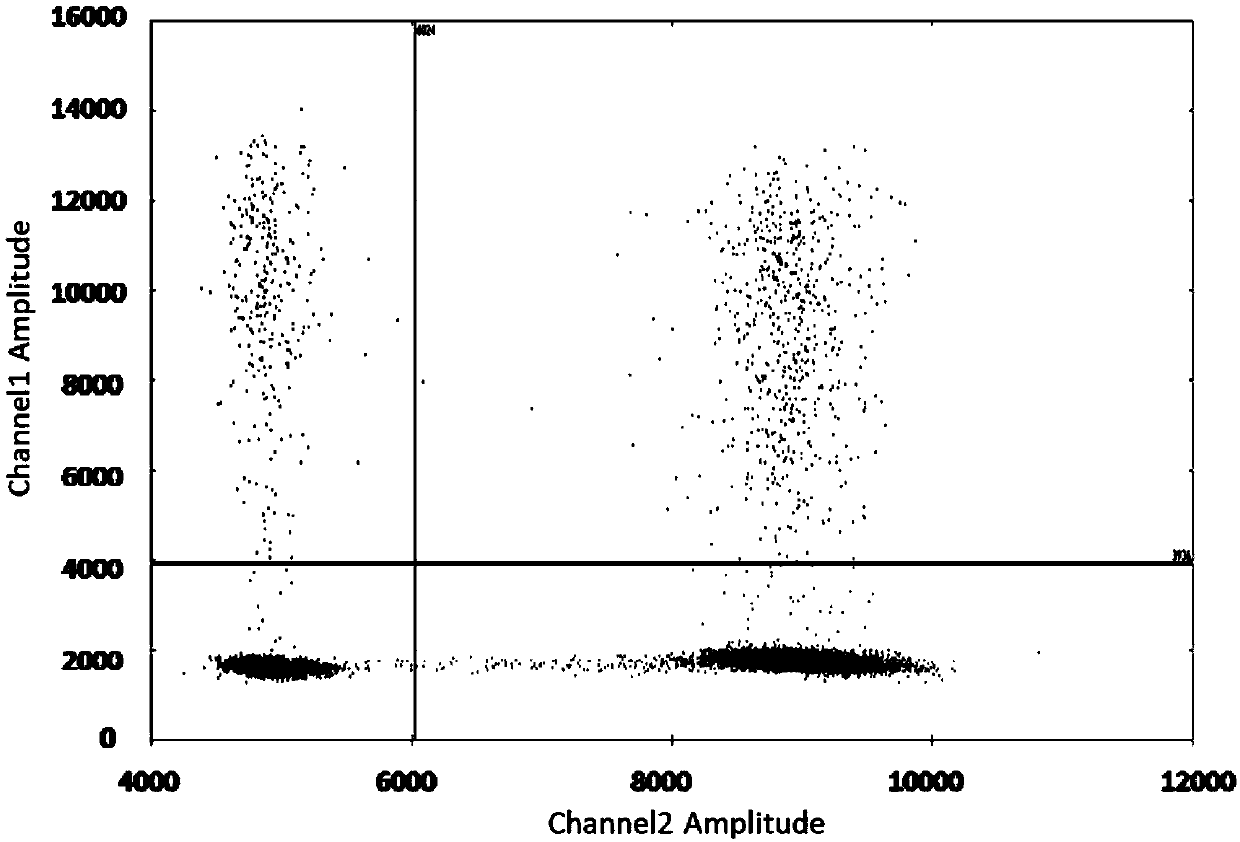
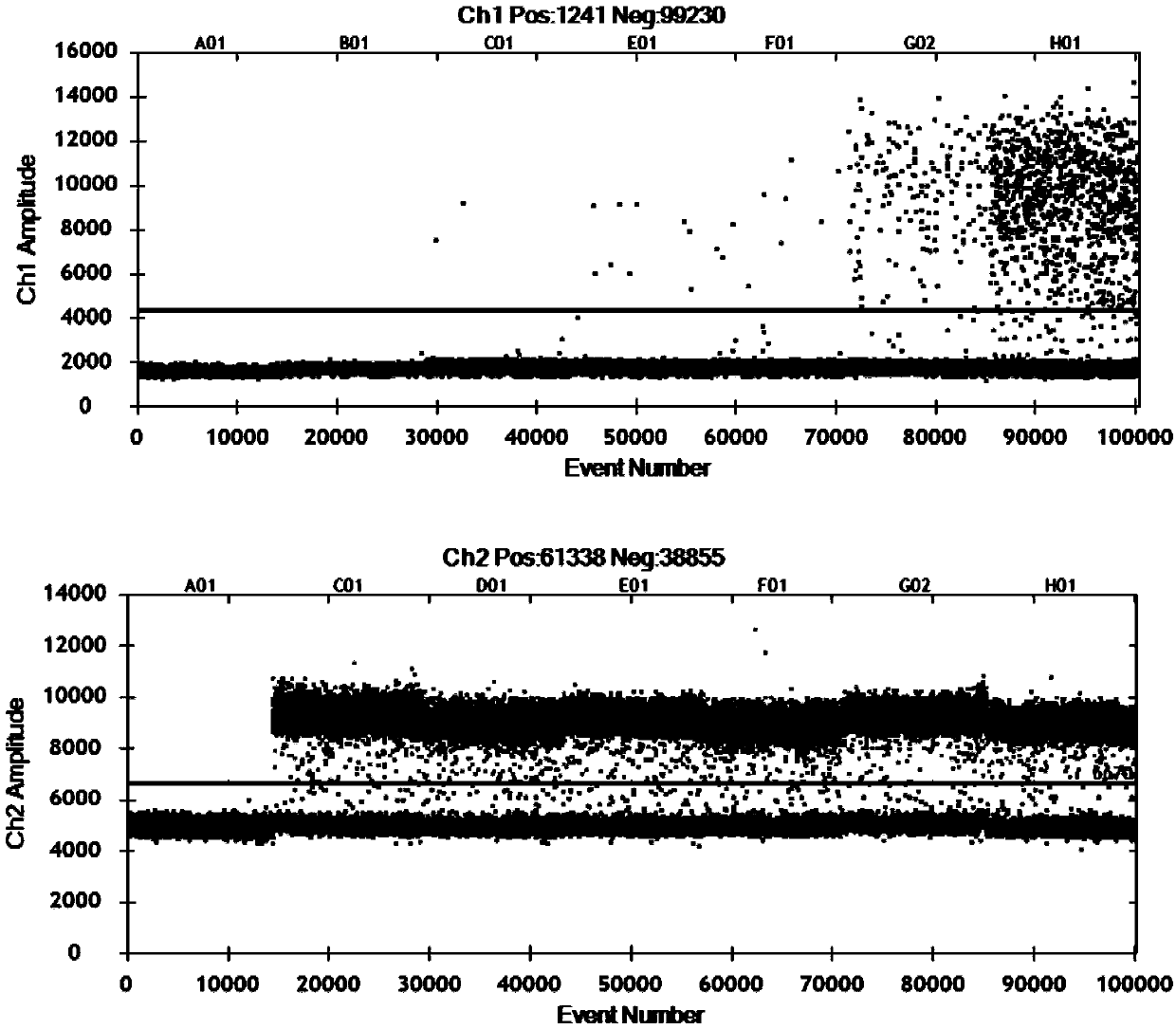
[0025] Example 1 Detection of EGFR gene T790M mutation site
[0026] The human EGFR gene mainly produces mutations in four exons, which are exons 18, 19, 20 and 21, of which exon 18 has 5 mutant types, exon 19 has 34 mutant types, and exon 20 has mutated types. There are 9 mutant types in exon 21 and 3 mutant types in exon 21. In this example, the most common exon 20 T790M mutation of the EGFR gene was selected to compare the ARMS-ddPCR method of the present application with the existing ARMS-qPCR method and ddPCR method.
[0027] Construction of EGFR Gene T790M Mutation Site Plasmid Standards
[0028] Site-directed mutation was performed on the T790M site by Overlap PCR method, and the mutant gene was constructed into a plasmid.
[0029] The primers used by the Overlap PCR method are exon 20 primers and T790M site primers, as shown in Table 1.
[0030] Table 1 Construction of EGFR gene T790M mutation site primer list
[0031]
[0032] The 20-exon primers F and T790M R,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com