Screening method of solo-LTR toxic candidate strain with Inago2 in AvrPiz-t
An avrpiz-t, 1.avrpiz-t technology, applied in the field of molecular biology, can solve the problems of screening and identification primers and methods that do not retrieve virulent strains
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
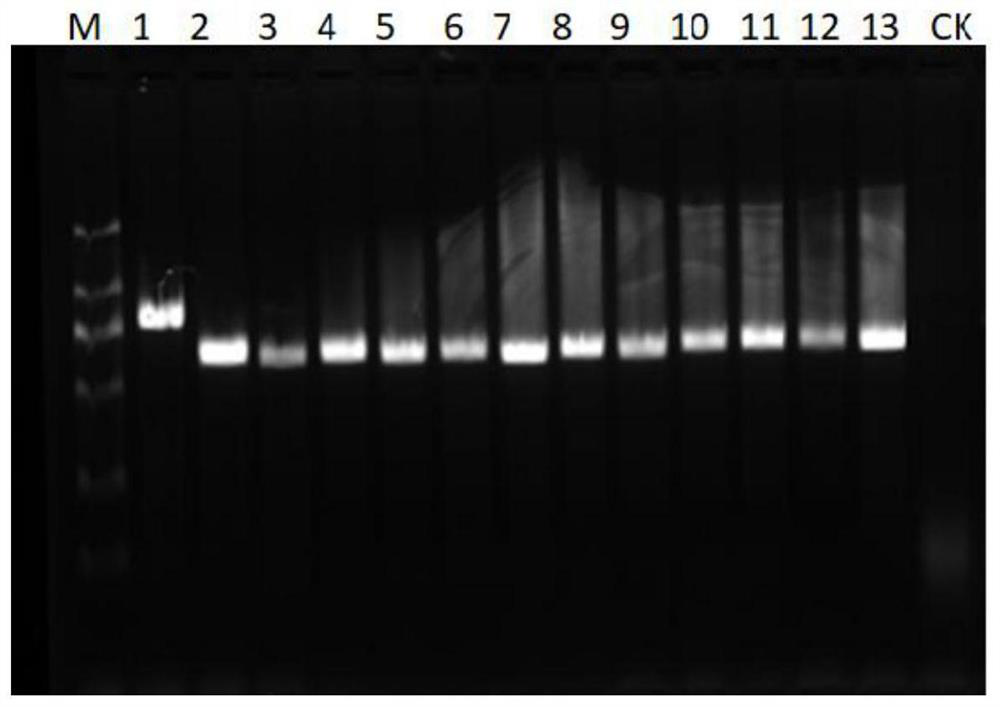
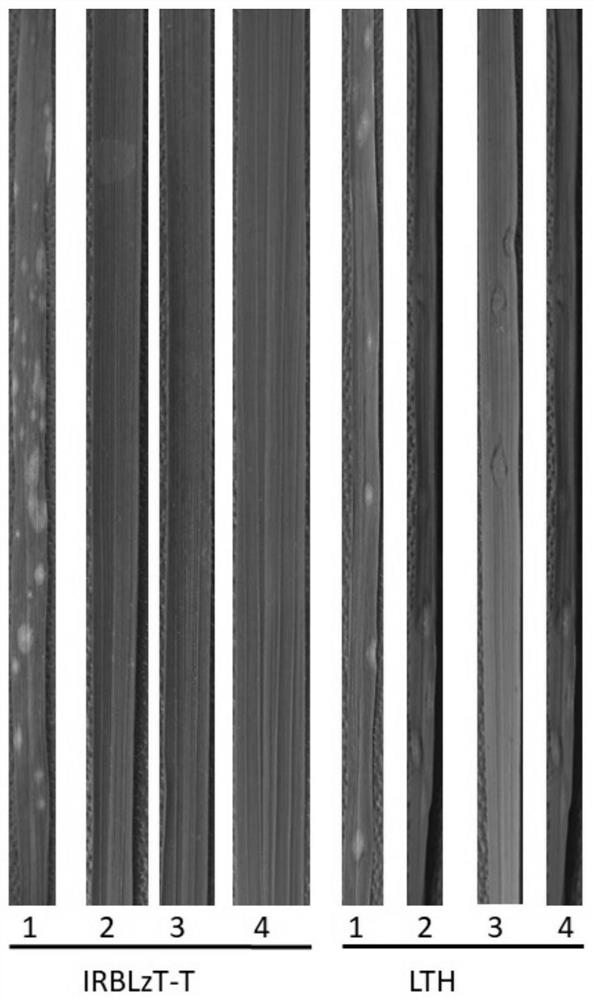
[0030] 1. Steps and verification of specific primer design
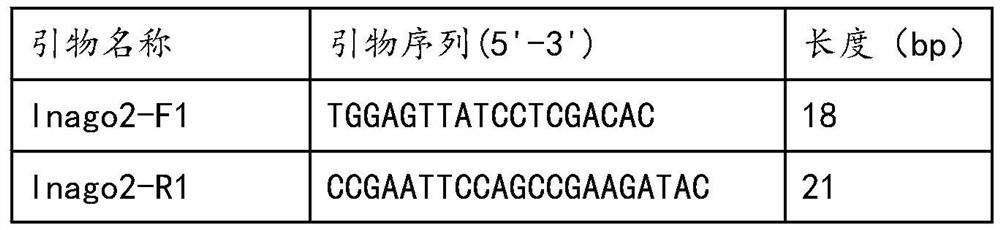
[0031] Primer design: compare the sequence of the upstream region of the insertion site of the AvrPiz-t gene promoter between the rice blast strains containing the insertion and those without the insertion, and select a short oligonucleotide sequence (with a GC content of about 50%) in the conserved region as the front primer; Then a conserved oligonucleotide sequence (with a GC content of about 50%) is selected downstream of the gene coding region (CDS), and its reverse complementary sequence is used as a back primer. Finally, compare whether the primer sequences before and after are easy to form primer dimers, and those that are easy to form primer dimers are redesigned. The positions of the designed front and rear primers include the upstream promoter region and CDS region of the insertion site. Verification: Submit the designed primer sequence to the sequencing company for synthesis, and then use the synthesized...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com