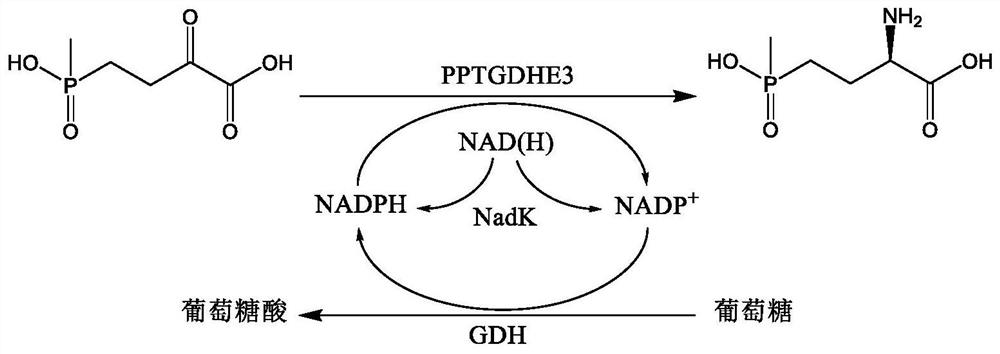
Coenzyme self-sufficient Escherichia coli as well as construction method and application thereof
A technology of Escherichia coli and coenzyme, which is applied in the application field of catalyzing and synthesizing glufosinate-ammonium, can solve problems such as insufficient concentration of coenzyme, and achieve the effects of shortening the reaction process, slowing down the short half-life and improving the space-time yield.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1: Knockout of Coenzyme Decomposition Gene
[0049] Single or double knockout of mudC, mazG and nadR on Escherichia coli BL21 (DE3), wherein the gene knockout plasmid used in the present invention is pCasRed, pCRISPR-gDNA (sgRNA) and homology arm (donor) into Escherichia On coli BL21(DE3), Cas9 / sgRNA induces a double-strand break at the target gene locus, and the recombinase Red integrates the donor into the target gene to achieve gene knockout, which is verified by sequencing. mudC sgRNA, mudCdonor, mazG sgRNA, mazG donor, nadR sgRNA, and nadR donor are respectively shown in SEQ ID NO 16 to SEQ ID NO 21 in the sequence table. The changes in intracellular coenzyme concentration of the strains obtained by different knockout combinations are shown in Table 1.
[0050] Table 1: Coenzyme changes after gene knockout
[0051] strain NADP(H) coenzyme concentration (μmol / g) E.coli BL21(DE3) 21.4 E.coli BL21(DE3)ΔmudC 24.7 E.coli BL21(DE3)Δ...
Embodiment 2
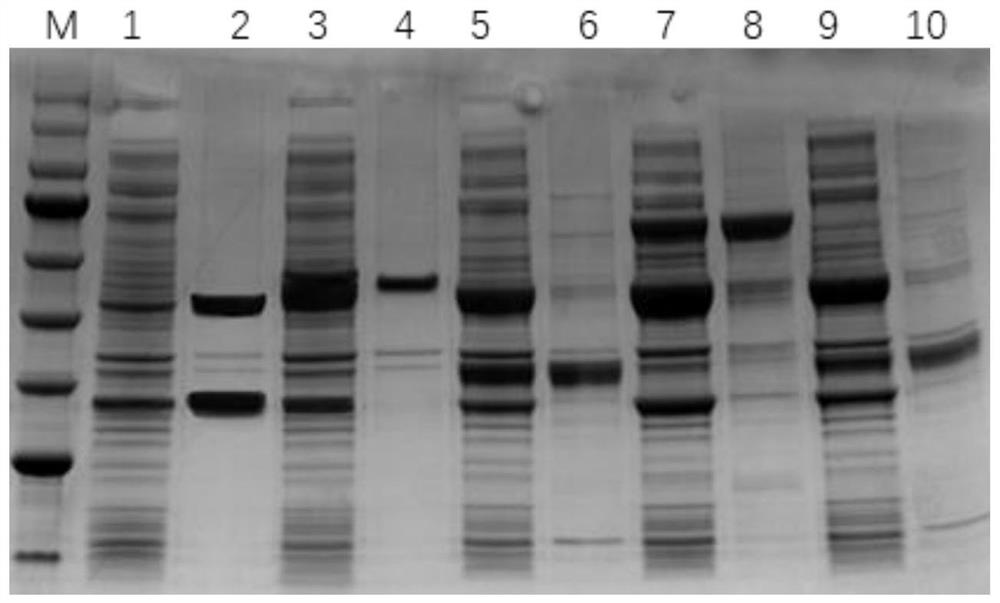
[0053] Example 2: Construction of Escherichia coli comprising NADH kinase
[0054] In this example, five NADH kinase sequences were selected from the NCBI database through literature reports, which were derived from Corynebacterium glutamicum (CgNadK) (accession number NC_003450.3), Escherichia coli (EcNadK) (accession number NC_000913.3), Methanocaldococcus jannaschii (MjNadK) (accession number NC_000009.12), Entamoeba histolytica (EhNadK) (accession number NW_001914860.1), Saccharomycescerevisiae (ScNadK) (accession number NC_001148.4), and the whole gene synthesis after codon optimization, nucleoside The acid sequences are respectively shown in SEQ ID NO.1-NO.5.
[0055] MjNadK, EcNadK, CgNadK, EhNadK, ScNadK were respectively cloned into the first multiple cloning site of plasmid pCDFDuet-1 by one-step cloning method:
[0056] Design carrier linearization primer 1 and primer 2, with the beginning and end of the linearized vector each 10-15 bp as homologous sequences, design...
Embodiment 3
[0089] Example 3: Construction of co-expression Escherichia coli
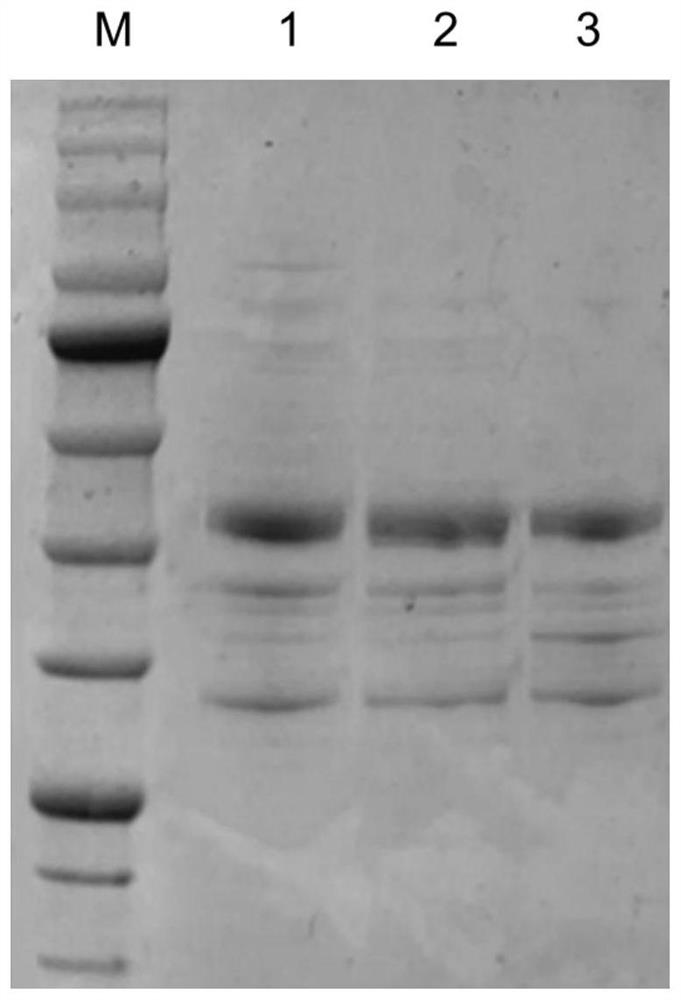
[0090]The recombinant Escherichia coli E. coli BL21(DE3) / pETDuet-1-PPTGDHE3-GDH containing glutamate dehydrogenase and glucose dehydrogenase has been constructed in the previous work of our laboratory (patent publication number CN109609475A). E.coli BL21(DE3) / pETDuet-1-PPTGDHE3-GDH, E.coli BL21(DE3)MN / pCDFDuet-1-MjNadK, E.coli BL21(DE3)MN / pCDFDuet-1-EcNadK, E.coli BL21(DE3)MN / pCDFDuet-1-CgNadK, E.coli BL21(DE3)MN / pCDFDuet-1-EhNadK, E.coli BL21(DE3)MN / pCDFDuet-1-ScNadK, E.coli BL21(DE3)MN / pCDFDuet-1-nadE, E.coli BL21(DE3)MN / pCDFDuet-1-nadD, E.coli BL21(DE3)MN / pCDFDuet-1-pncB were respectively inoculated into LB liquid medium, cultured for 12 hours and then extracted Plasmids, pETDuet-1-PPTGDHE3-GDH, pCDFDuet-1-MjNadK, pCDFDuet-1-EcNadK, pCDFDuet-1-CgNadK, pCDFDuet-1-EhNadK, pCDFDuet-1-ScNadK, pCDFDuet-1-nadE pCDFDuet- 1-nadD, pCDFDuet-1-pncB, measure the nucleic acid concentration, take 200ng pETDuet-1-PPTGDH...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com