Code word design method for DNA storage
A design method and DNA sequence technology, applied in bioinformatics, instruments, etc., can solve the problems of high synthesis cost and low coding rate, and achieve the effects of simple structure, improved coding rate and reduced cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
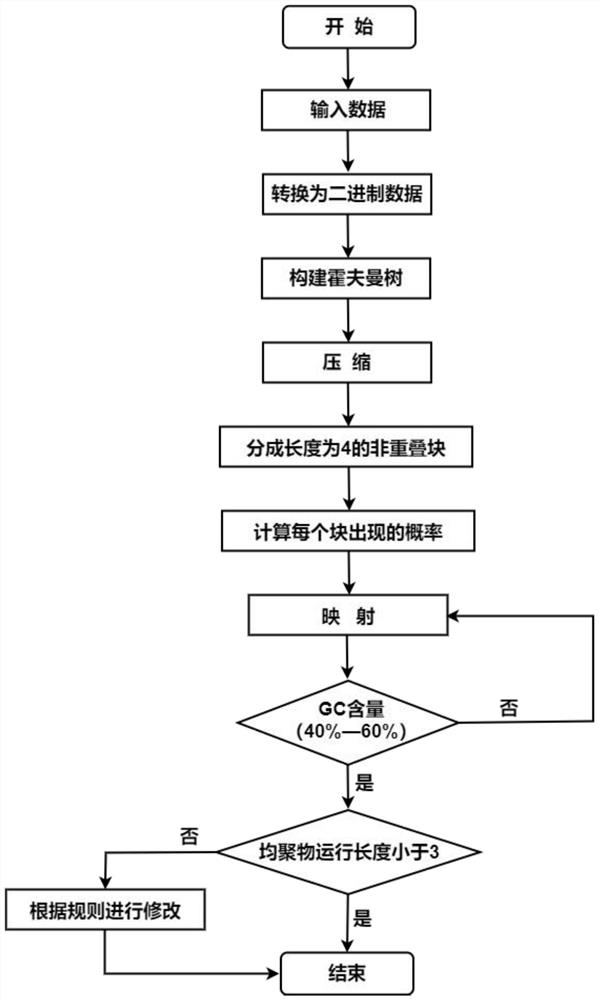
[0040] The embodiments of the present invention are implemented on the premise of the technical solutions of the present invention, and detailed implementation methods and specific operation processes are given, but the protection scope of the present invention is not limited to the following embodiments. In the embodiment, this encoding algorithm is used to encode a text file with a size of 511B, and the constraints satisfied are as described above.
[0041] Step 1: Take input data and convert to binary data.
[0042] Specifically, first use the abs function to convert the characters in the text file into ASCII codes, and then convert them into 8-bit binary numbers. The 511B text file can be converted into 4088bits binary data through the above operations.
[0043] Step 2: Compress the binary data.
[0044] It should be noted that each 16-bit binary number is taken as a group, and then compressed using the minimum variance Huffman tree, where the Huffman tree is constructed ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com