Methods, devices, arrays and kits for detecting and analyzing biomolecules
a biomolecule and array technology, applied in the field of biomolecule detection and analysis methods, can solve the problems of low biomolecule specificity and high affinity of certain membranes used in such methods, and achieve the effect of increasing specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
first embodiment
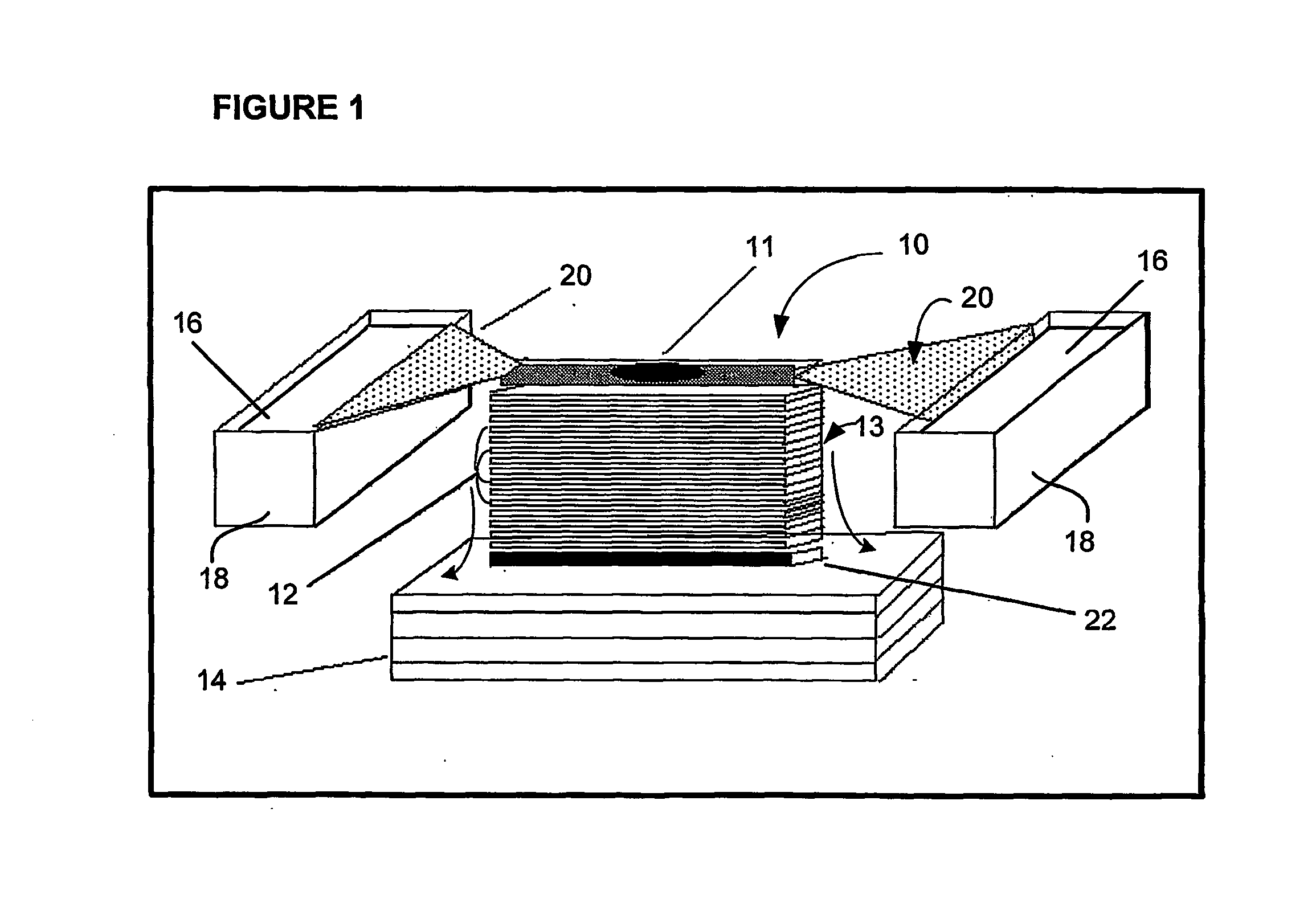
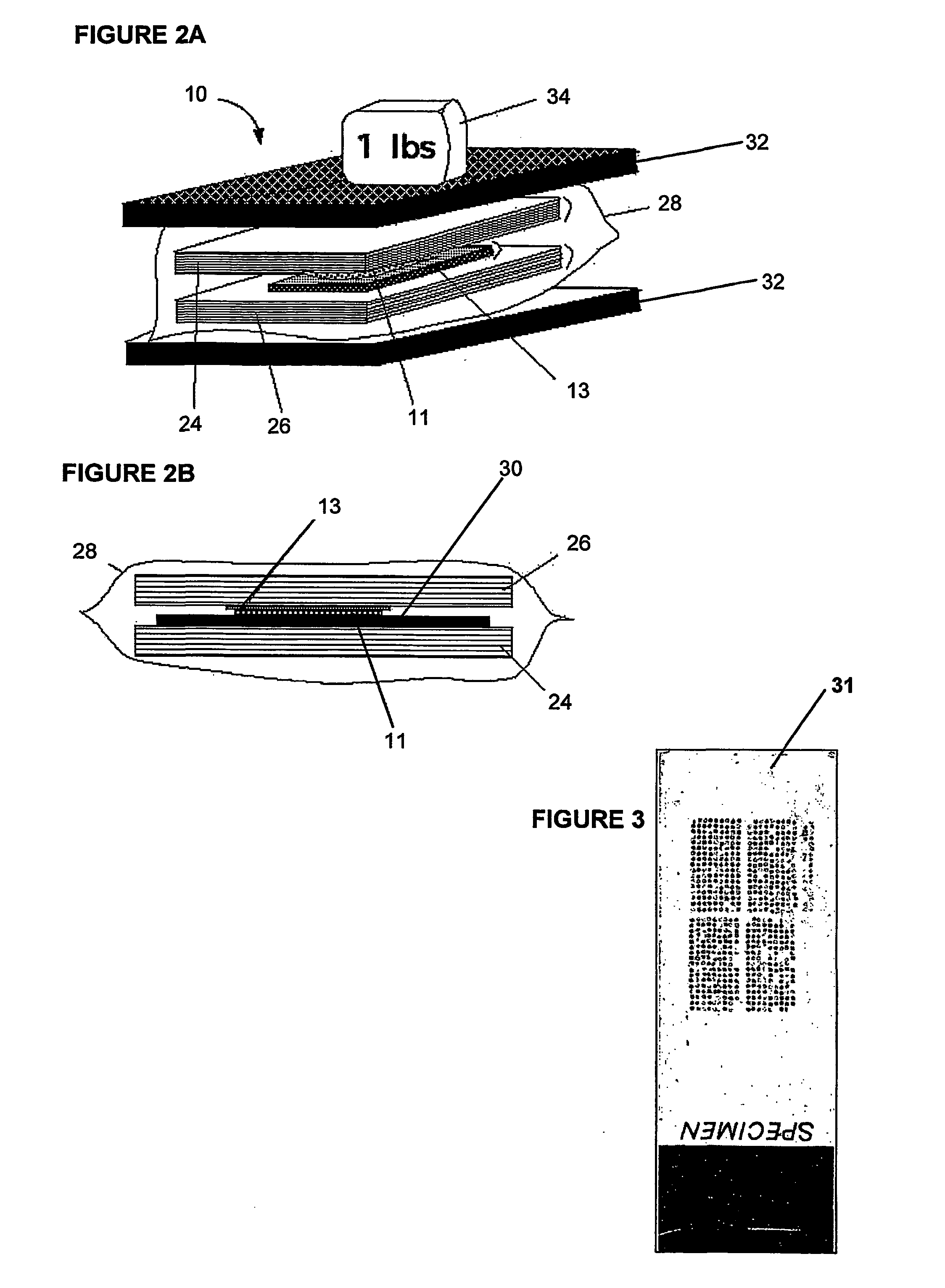
[0155] More specifically, in a first embodiment, membrane stack 13 comprises one or more membranes 12, for instance up to five membranes, generally constructed as described herein. The membranes 12 in stack 13 should have a high affinity for proteins and other biomolecules but have a low capacity for retaining such molecules. This feature permits the molecules to pass through the membrane stack with only a limited number being trapped on each of the successive layers, thereby allowing multiple "carbon copies" (substantial copies that are not necessarily identical copies, they may have slight differences but can be identical or nearly identical) to be generated. In other words, the low capacity allows the creation of multiple replicates as only a limited quantity of the biomolecules are trapped on each layer.
[0156] First and second filter pads 24, 26 are preferably constructed of a blotting paper such as GB004 Blotter Paper available from Schleicher and Schuell. Filter pads 24, 26 ar...
second embodiment
[0230] Thus, with another embodiment, each of the membranes will bind to a different group of proteins essentially permitting "3-D gel electrophoresis" by allowing proteins to be separated into three dimensions: in the X and Y dimensions by charge and mass, and then in the Z dimension by an additional chemical characteristic. The proteins on the membranes can be visualized by the immuno-staining and imaging methods set forth below. They may also be advantageously analyzed by mass spectrometry either without additional cleavage or after such cleavage (see, e.g., WO00 045168), or by other means. Examples of the methods and kits facilitate such analysis because the stratification by the different membranes helps to expose moderate and low abundance protein spots that would otherwise be undetectable on standard 2-D gels. The more spots that are available for analysis, the more data can be generated by mass spectroscopy or by such other approaches.
[0231] Other Membrane Characteristics
[0...
example detection
[0257] Example Detection Chemistries with Detector Cocktails
[0258] In certain embodiments, after proteins have been transferred through the membrane stack, individual membranes layers are separated and each is incubated in a separate antibody (or other detector molecule) cocktail. A key advantage of creating multiple replicate blots is that many more detector molecules (e.g., antibodies) can be usefully employed than if all of the detectors had to be crowded onto a single blot.
[0259] An exemplary process for designing the ligand cocktails--and for determining which proteins will be identified on each membrane layer--is provided below. First the panel of proteins of interest is selected. These can be randomly selected proteins and / or proteins that are not directly related to one another or may be groups of known proteins previously implicated to play a role in one or more particular cellular phenomena (e.g. apoptosis, cell cycle progression) or a particular disease (e.g. prostate can...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
| thick | aaaaa | aaaaa |
| thick | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com