Preservation of RNA in a biological sample
a biological sample and rna technology, applied in the field of preserving rna in biological samples, can solve the problems of limiting the information available from analysis of biological samples, and affecting the quality of biological samples,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
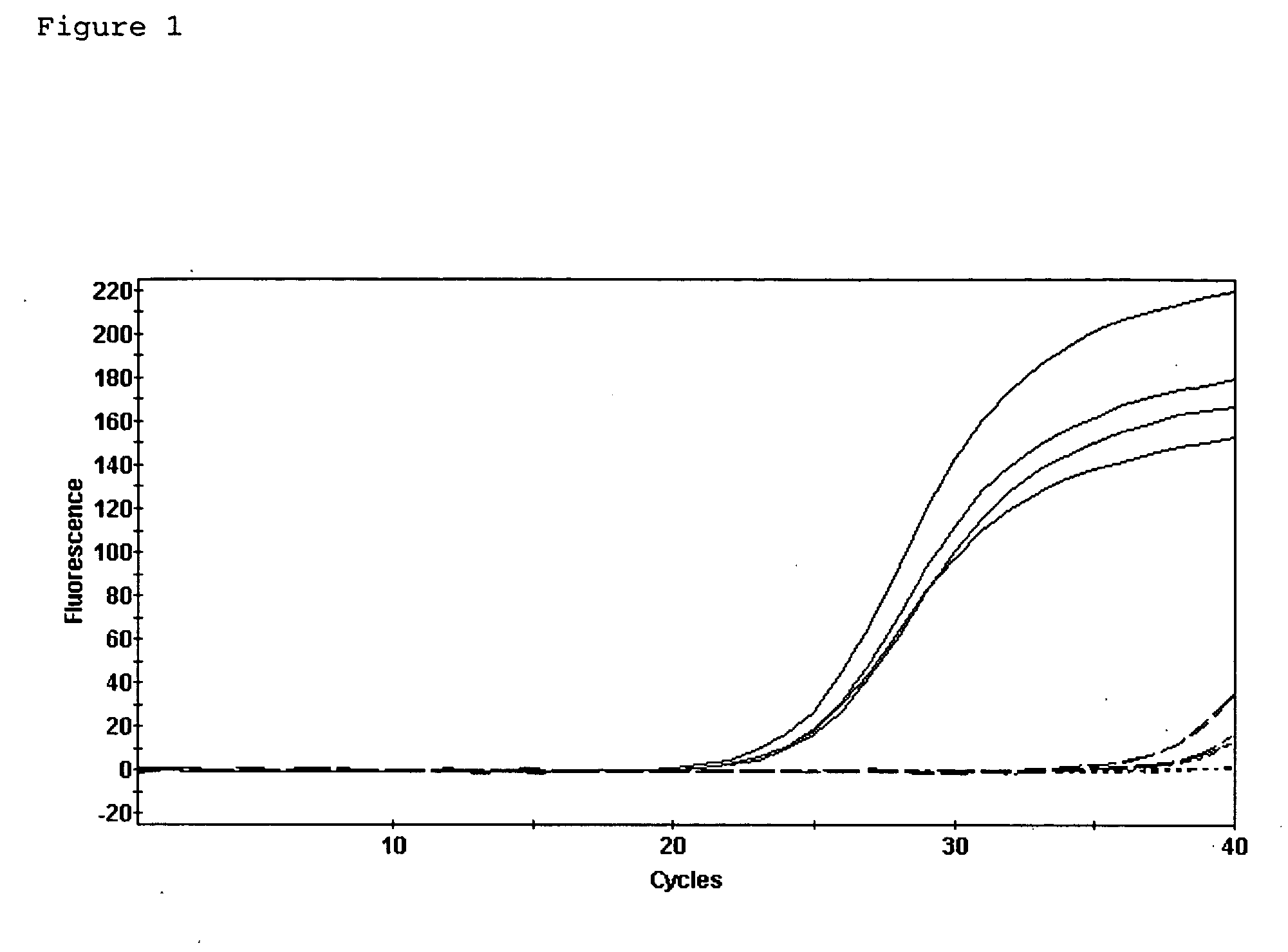
RNA Diffusion Into the Tissue Incubation Buffer During Immunostaining
[0073] To characterize the mechanism by which RNA is lost from the tissue sections during immunostaining, RNA from immunostained tissue sections was extracted and compared to that from tissue sections that had not undergone immunostaining. Frozen sections were air-dried and fixed in cold acetone for 2 min. After a quick rinse in phosphate buffered saline (0.137 M NaCl, 0.0027 M KCl, 0.01 M phosphate buffer pH 7.4, PBS), sections were incubated in methyl green solution (Vector, Burlingame, Calif.) for 2 min. They were then rinsed in PBS and incubated in OX-42 antibody (Serotec, Raleigh, N.C.) diluted 1:100 in PBS with 0.5% acetylated BSA (Sigma) for 5 min. Sections were rinsed in PBS and then incubated in biotinylated goat anti-mouse antibody (Chemicon, Temecula, Calif.) diluted 1:100 in PBS with 0.5% acetylated BSA for 5 min. After rinsing in PBS, sections were incubated in 0.1 M Tris-HCl, pH 8.0 for 1 min and the...
example 2
Identification of RNA Preservatives
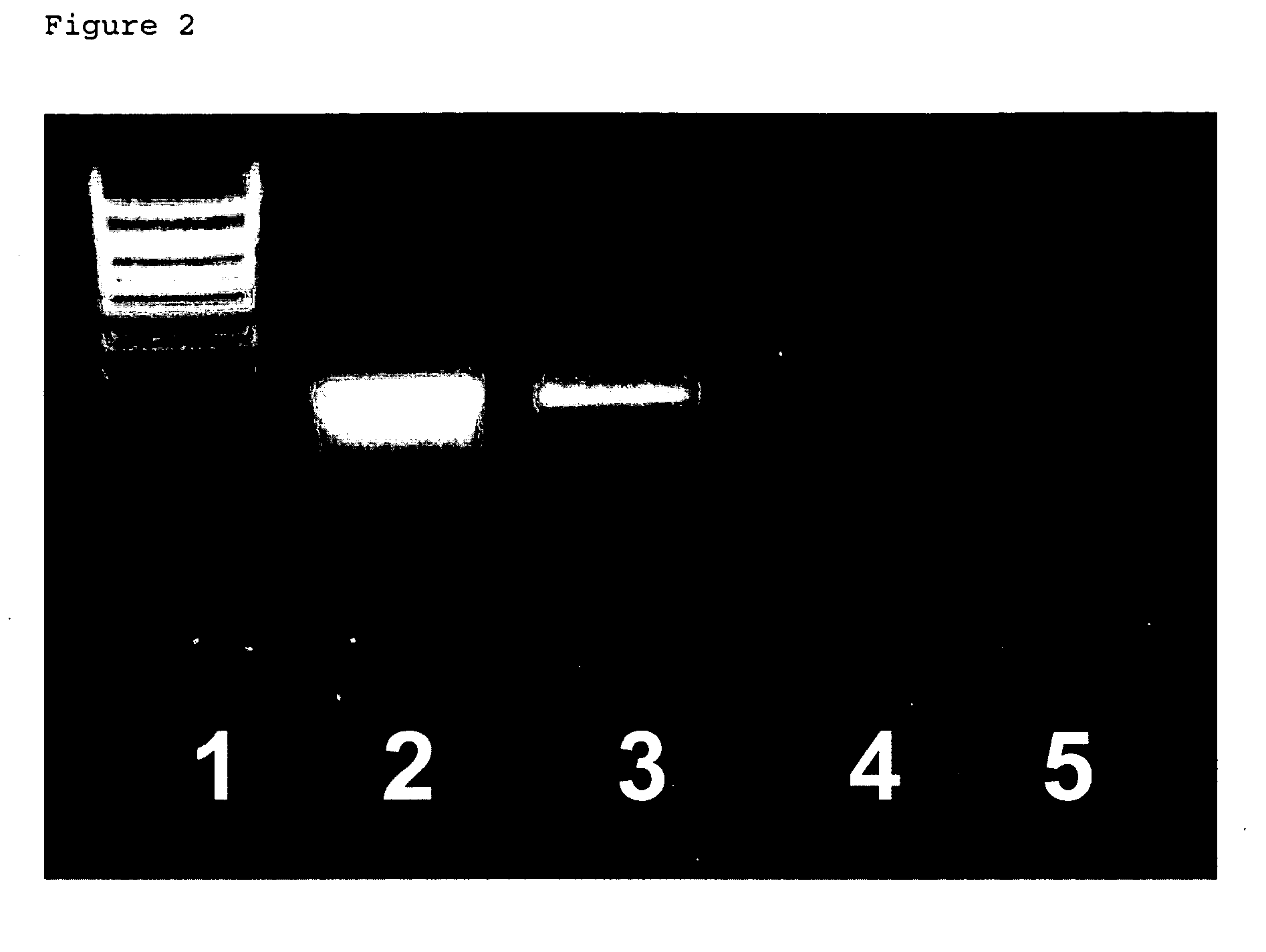
[0079] A testing compound, tris(4-aminophenyl)methane, was first incubated with RNA molecules in an aqueous solution, such as water. The mixture of the compound and RNA was then centrifuged at 10,000 g for 20 min to collect any precipitate. The collected precipitate was then analyzed by denaturing agarose gel electrophoresis. A band of the proper molecular weight (depending on the type of RNA used) indicated that the compound precipitated RNA.
[0080] Other compounds that precipitated RNA from an aqueous buffer included methyl green, cresyl violet, tris(4-aminophenyl)methane and hexamine cobalt. Compounds that were found positive by this screening assay were then tested for efficacy in preventing RNA loss during immunohistochemistry on brain tissue sections.
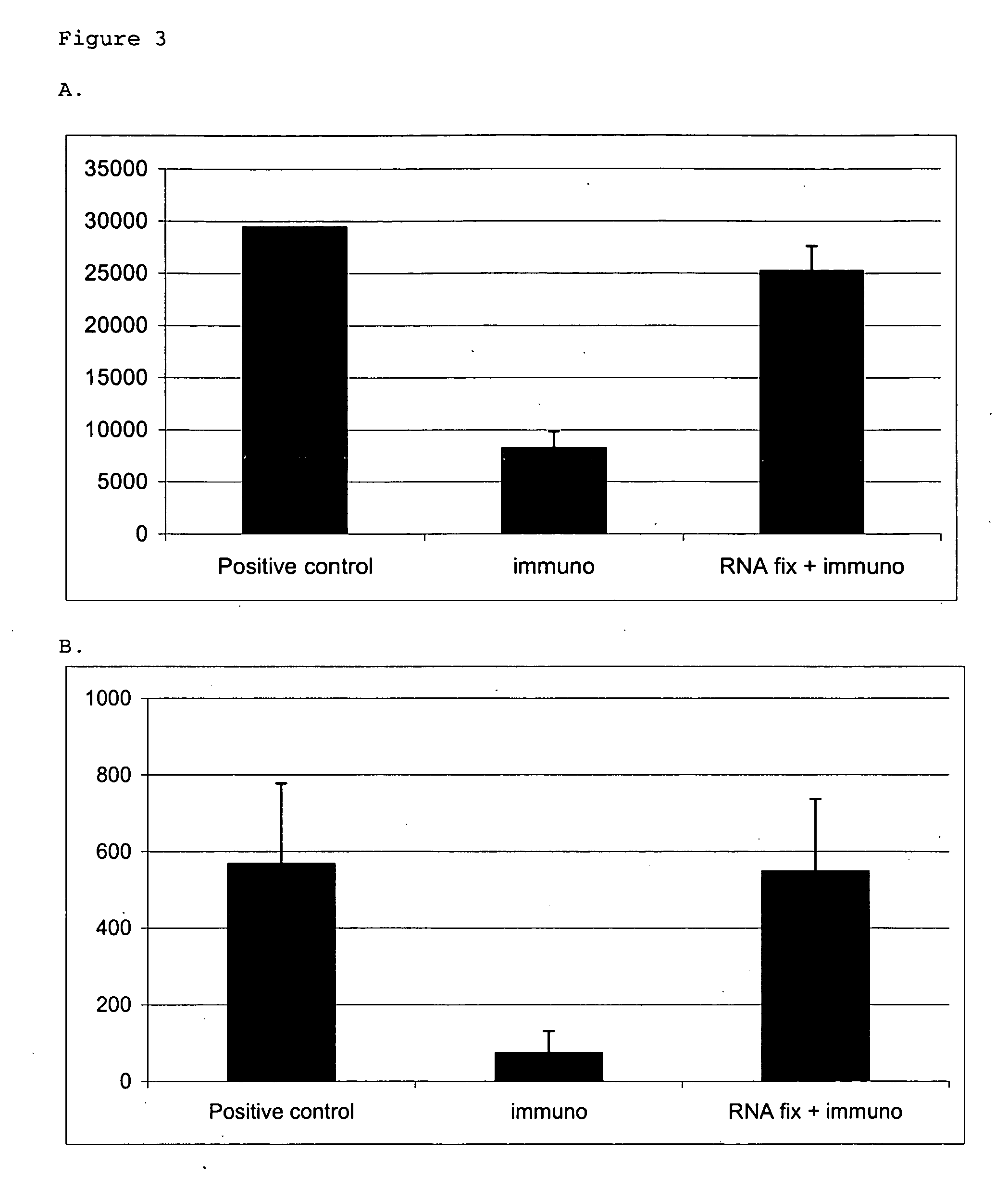
example 3
Immunohistochemical Assay
(A) Comparison Example—OX42 Immunostaining Without RNA Preservation
[0081] Frozen sections of tissues were air dried and fixed in cold acetone for 2 min. Sections were rinsed in PBS and incubated with OX42 antibody (Serotec, Raleigh, NC) diluted 1:100 in PBS with 0.5% acetylated BSA (Sigma) for 5 min. Sections were then rinsed in PBS and incubated with biotinylated goat-anti-mouse antibody (Chemicon, Temecula, Calif.), diluted 1:100 in PBS with 0.5% acetylated BSA for 5 min. Slides were again rinsed in PBS and then incubated 1 min in 100 mM Tris-HCl, pH 8. Then, slides were incubated with peroxidase-conjugated streptavidin (Jackson ImmunoResearch Laboratories, West Grove, Pa.), which was diluted 1:100 in PBS, for 5 min. Sections were rinsed in PBS, and detection was performed using AEC (DAKO, Carpinteria, Calif.) for 5 min. Sections were rinsed in water and counterstained with Mayer's Hematoxylin (BioGenex, San Ramon, Calif.) for 5 sec. Sections were rinsed...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com