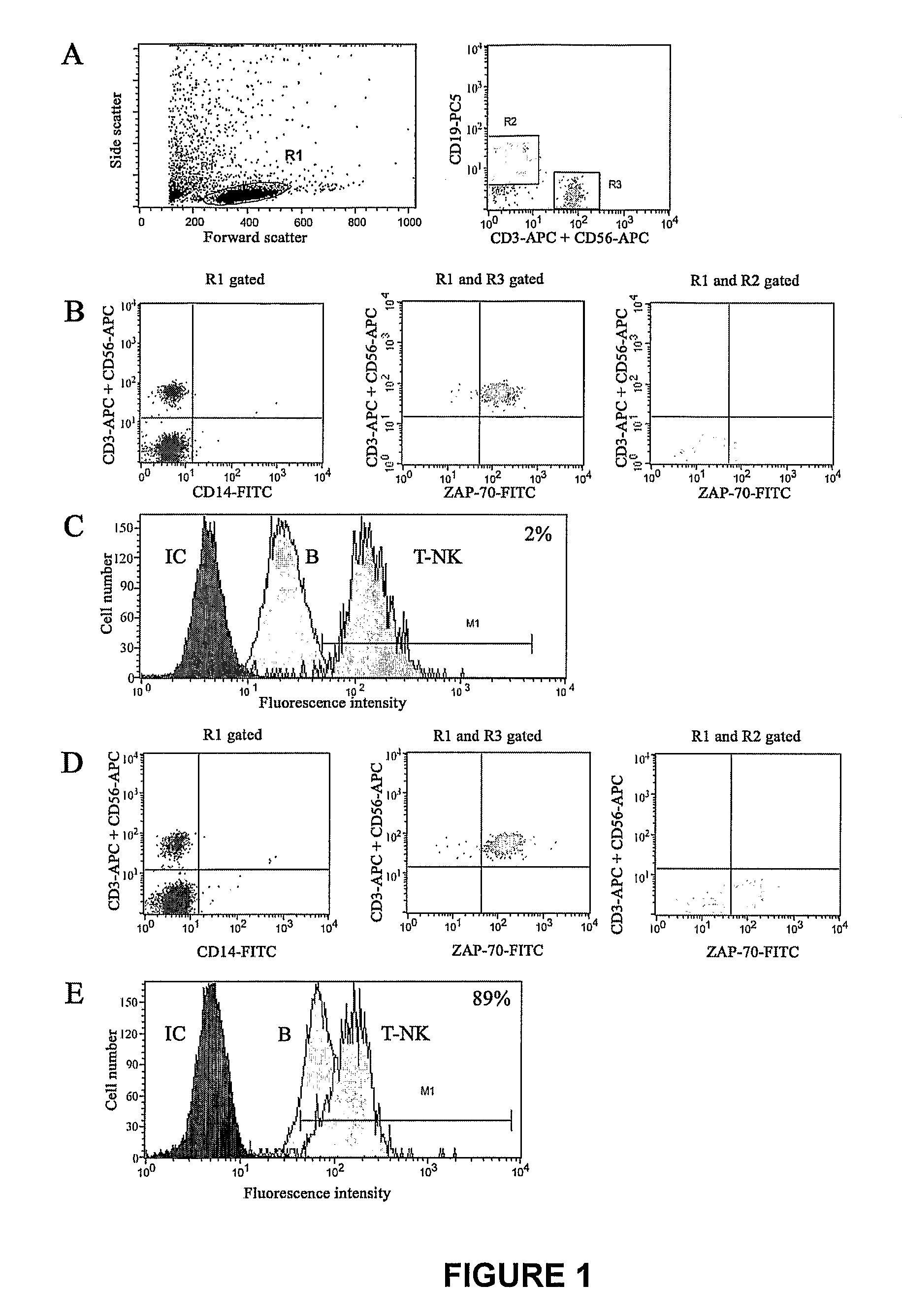
Method of Diagnosis/Prognosis of Human Chronic Lymphocytic Leukemia Comprising the Profiling of Lpl/Adam Genes
a technology of human chronic lymphocytic leukemia and lpl/adam gene, which is applied in the direction of microbiological testing/measurement, biochemistry apparatus and processes, etc., can solve the problems of inaccessibility to most medical facilities, high cost of analysis, time-consuming, etc., and achieve accurate estimation of the prognosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Patients' Characteristics
[0160]Table 2 summarizes the 127 patients' characteristics:
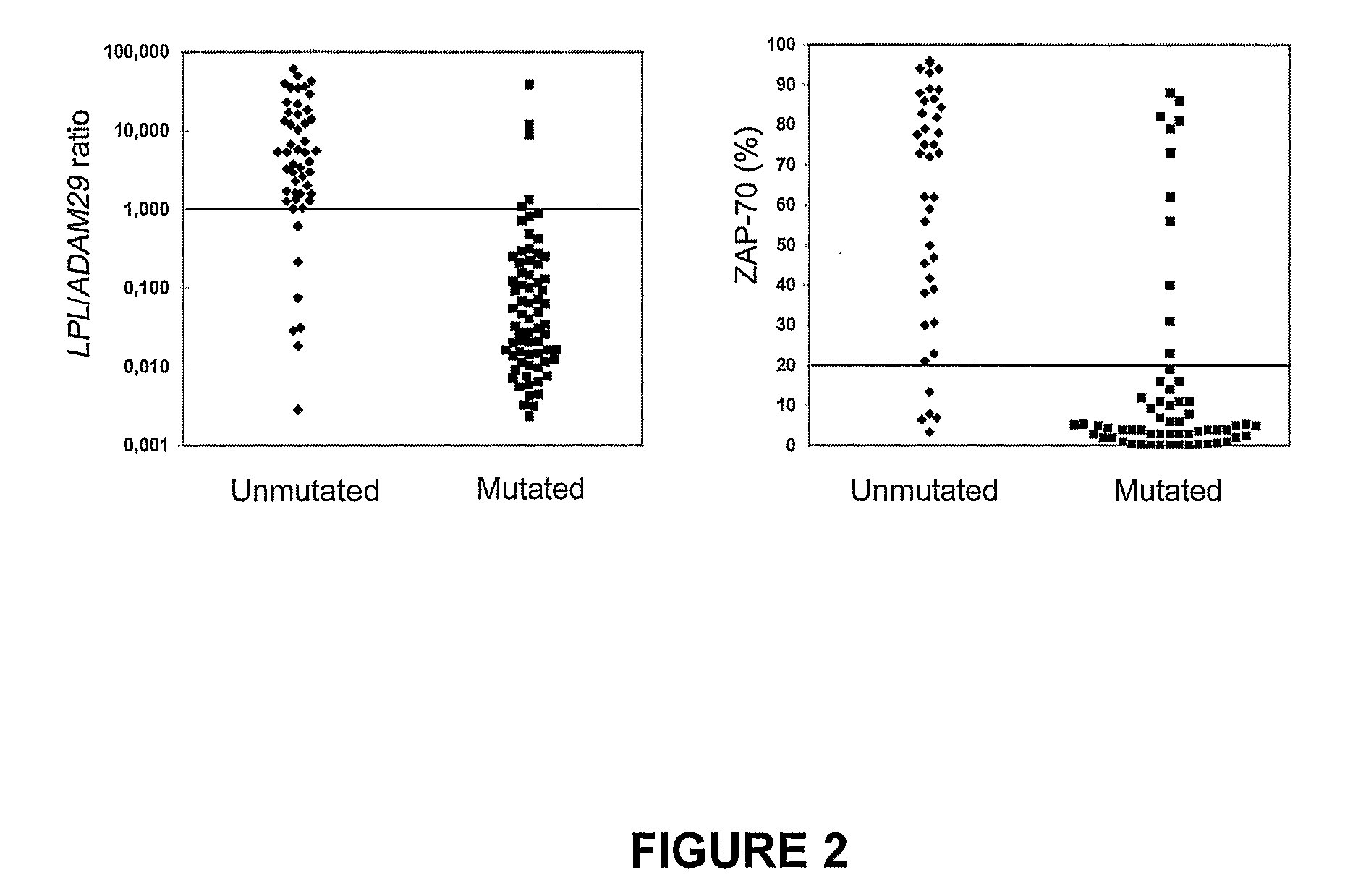
TABLE 2Clinical and biological characteristics of patientsStagesStagesStage AStage BStage CB + CA + B + CNo. Patients87291140127Age, years*63.058.059.058.561.0Male sex51 (59%)24 (83%) 5 (45%)29 (72%)80 (63%)Lymphocyte count, × 109 / L*15.237.2151.845.019.0Hemoglobin, g / dL*14.113.47.213.013.8Platelets, × 109 / L*212.5160.0187.5172.0198.0Lymphocyte doubling time>12 months64 (84%)NANANANA12 (16%)NANANANAIgVH genesUM27 (31%)17 (59%) 9 (82%)26 (65%)53 (42%)MT60 (69%)12 (41%) 2 (18%)14 (35%)74 (58%)ZAP-7045 (65%) 7 (30%) 3 (33%)10 (31%)55 (54%)≧20%24 (35%)16 (70%) 6 (67%)22 (69%)46 (46%)L / A ratio56 (69%)13 (46%) 2 (20%)15 (39%)71 (60%)≧125 (31%)15 (54%) 8 (80%)23 (61%)48 (40%)Progression22 (25%)NANANANACLL-related death4 (5%)11 (38%) 6 (55%)16 (40%)20 (18%)*Median valuesUM indicates unmutated;MT, mutated;L / A, LPL / ADAM29;NA, not applicable.
[0161]Fifty three patients (42%) were allocated to the UM IgVH group whi...
example 2
LPL and ADAM29 are Better Surrogate Markers for IgVH Mutational Status than SPG20 and NRIP1
[0162]LPL, SPG20, ADAM29 and NRIP1 were first evaluated in an initial series of 71 patients. The present inventors quantified the expression of these 4 genes on total lymphocytes, since preliminary experiments in five patients showed similar results as compared to purified leukemic cells (data not shown). For each gene, the present inventors determined which expression levels could best segregate UM from MT patients using Youden's and validity indexes. Results showed that overall LPL and ADAM29 performed better than SPG20 and NRIP1 to predict UM and MT IgVH profiles respectively (Table 3). The concordance rate was 83% for ADAM29, 77% for LPL, 65% for SPG20 and 45% for NRIP1.
TABLE 3Correlation of gene expression with IgVH mutational statusLPL / GeneLPLADAM29SPG20NRIP1ADAM29expression‡≧1≧3≧3.5≧4≧1UM (n = 37)30782921131716325MT (n = 34)9252867271915331Sensitivity*81%82%57%56%86%Specificity*74%78%79...
example 3
Reproducibility of LPL and ADAM29 Quantification
[0164]Since LPL and ADAM29 appeared to better reflect the IgVH mutational status, the present inventors evaluated the reproducibility of their quantification by RQ-PCR. This was done by comparing results obtained from replicate samples for 4 patients, 2 overexpressing LPL and 2 overexpressing ADAM29. For each patient, 4 replicates were analyzed during the same reaction run (intra-run variability), and this was repeated on 3 different days (inter-run variability). The overall variability of these 12 replicates is shown in Table 4. Of note, intra-run variability was always smaller than overall variability, with CV being less than 0.5% for LPL and less than 1.1% for ADAM29 (data not shown).
TABLE 4Reproducibility of RQ-PCRLPLADAM29PatientCt ± SDCV (%)Ct ± SDCV (%)CLL-7323.54 ± 0.461.97NANACLL-10523.96 ± 0.381.59NANACLL-67NANA27.33 ± 1.184.31CLL-101NANA23.322.21Ct indicates threshold cycle;SD, standard deviation;CV, coefficient of variation...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com