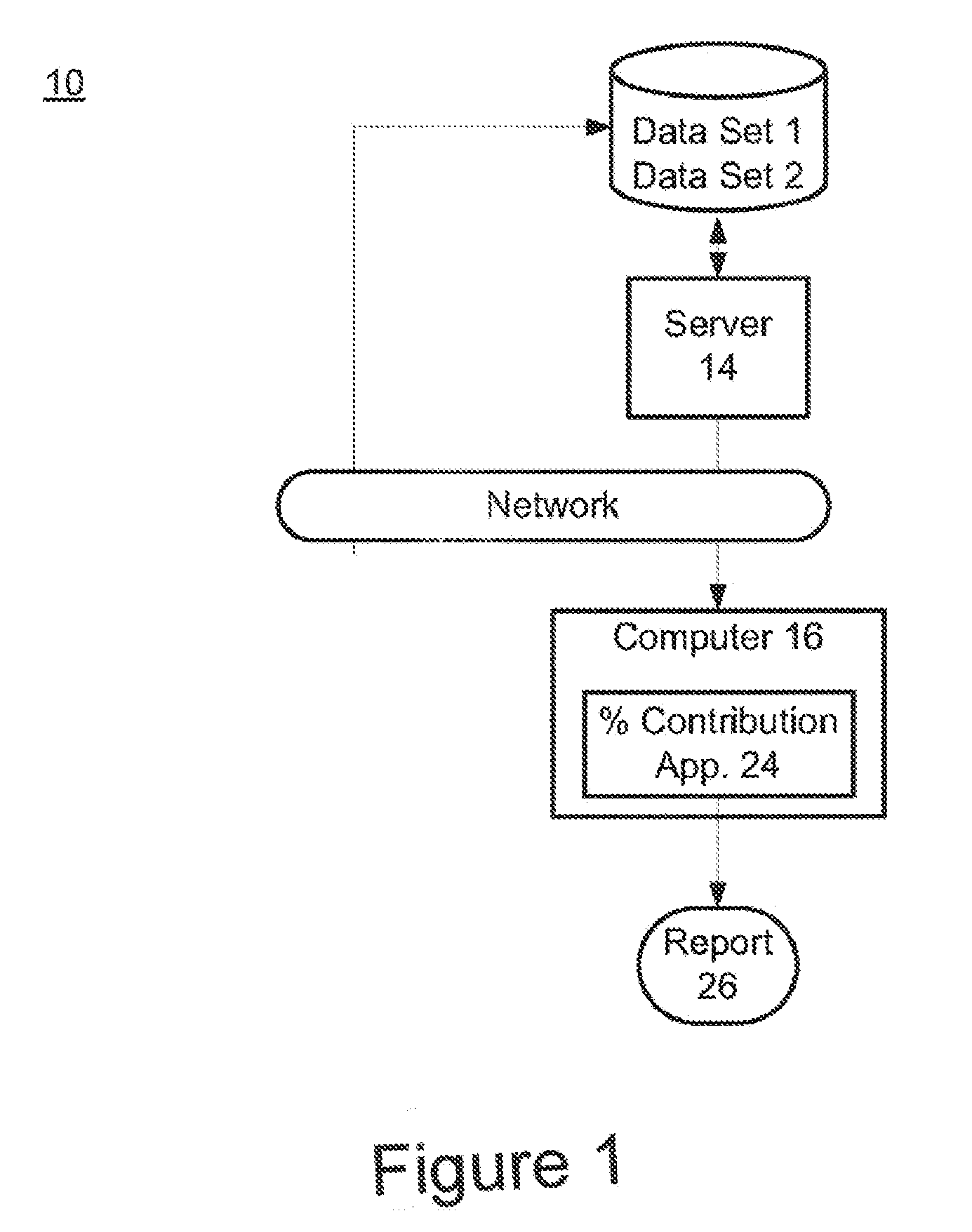
Risk calculation for evaluation of fetal aneuploidy
a risk calculation and fetal aneuploidy technology, applied in the field of noninvasive methods, can solve problems such as the inability of the system to reliably perform analysis, and achieve the effect of determining the risk of fetal aneuploidy for the first fetal chromosom
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Subjects
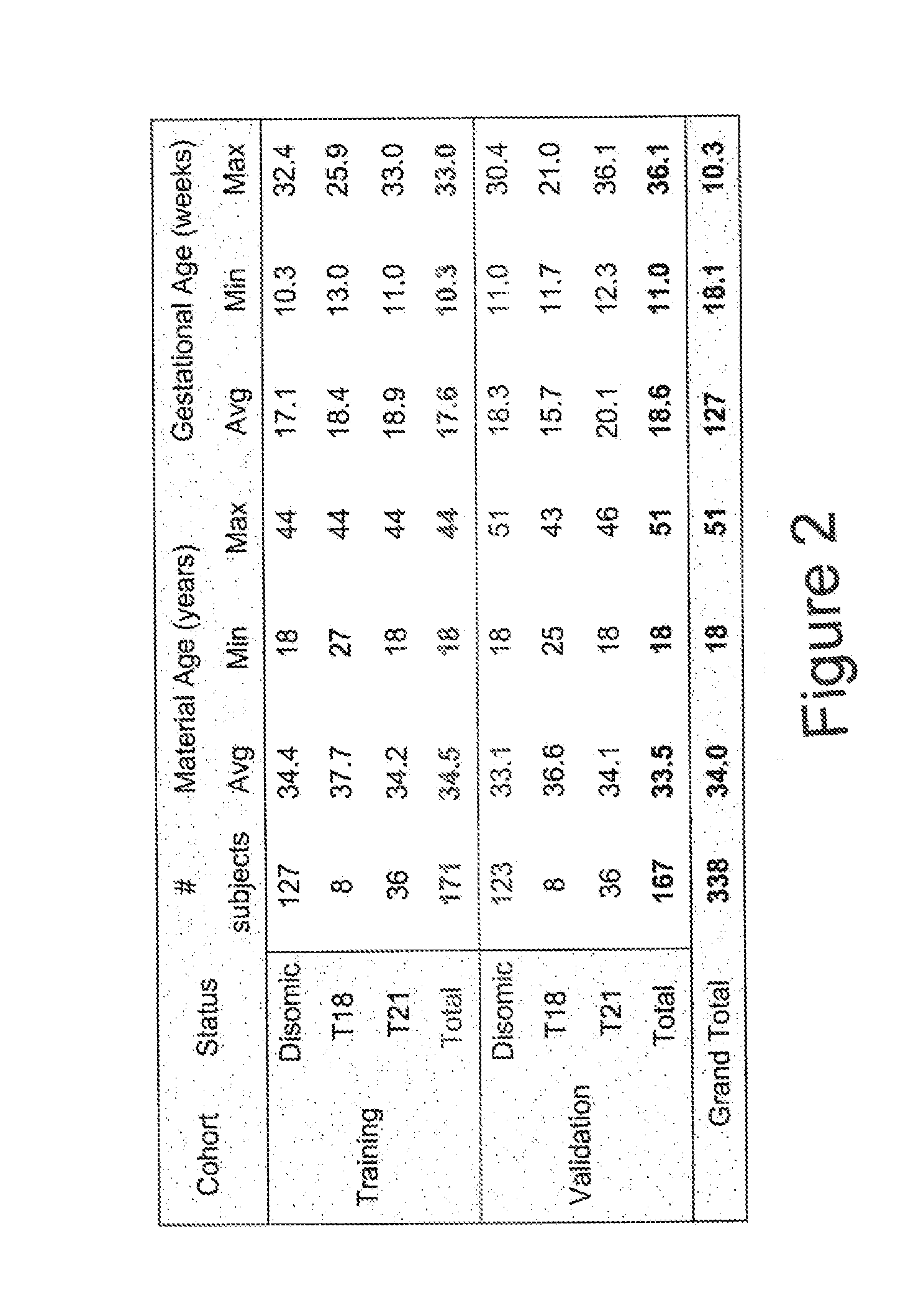
[0092]Subjects were prospectively enrolled upon providing informed consent under protocols approved by institutional review boards. Subjects were required to be at least 18 years of age, at least 10 weeks gestational age, and to have singleton pregnancies. A subset of enrolled subjects, consisting of 250 women with disomic pregnancies, 72 women with trisomy 21 (T21) pregnancies, and 16 women with trisomy 18 (T18) pregnancies, was selected for inclusion in this study. The subjects were randomized into a first cohort consisting of 127 disomic pregnancies, 36 T21 pregnancies, and 8 T18 pregnancies, and a second cohort consisting of 123 disomic pregnancies, 36 T21 pregnancies, and 8 T18 pregnancies. The trisomy status of each pregnancy was confirmed by invasive testing (fluorescent in-situ hybridization and / or karyotype analysis). The trisomy status of the first cohort was known at the time of analysis; in the second cohort, trisomy status was blinded until after risk calculatio...
example 2
Analysis of Non-Polymorphic Loci to Estimate Chromosome Dosage
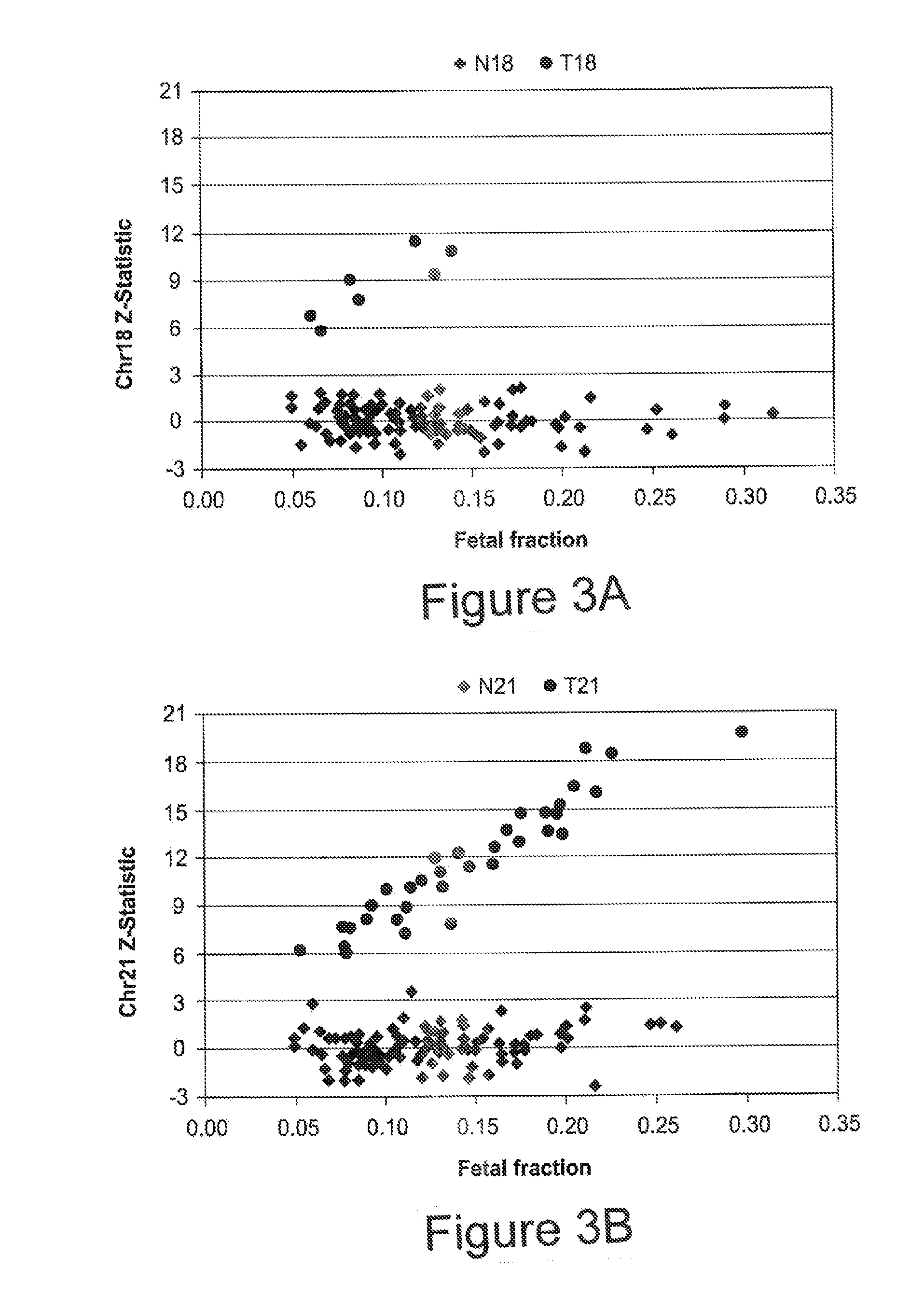
[0094]To estimate fetal chromosome dosage, assays were designed against 576 non-polymorphic loci on each of chromosome 18 and 21, where each assay consisted of three locus-specific oligonucleotides: a left oligo with a 5′ universal amplification tail, a 5′ phosphorylated middle oligo, and a 5′ phosphorylated right oligo with a 3′ universal amplification tail. The selected loci were used to compute a chr21 dosage metric and a chr18 dosage metric for each sample. First cohort samples were analyzed to identify 384 of the 576 loci on chr21 and chr18 best able to discriminate T21 and T18 from normal samples. First, sequence counts were normalized by systematically removing sample and assay biases using median polish (see Tukey, Exploratory Data Analysis (Addison-Wesley, Reading Mass., 1977) and Irzarry, et al., NAR, 31(4):e15 (2003)).
[0095]Next, the 384 loci on each chromosome exhibiting the greatest residual difference betwee...
example 3
Analysis of Polymorphic Loci to Assess Percent Fetal Contribution
[0097]To assess fetal nucleic acid proportion in the maternal samples, assays were designed against a set of 192 SNP-containing loci on chromosomes 1 through 12, where two middle oligos differing by one base were used to query each SNP. SNPs were optimized for minor allele frequency in the HapMap 3 dataset. Duan, et al., Bioinformation, 3(3):139-41 (2008); Epub 2008 Nov. 9.
[0098]Assays were designed against 576 non-polymorphic loci on each of chr18 and chr21, where each assay consisted of three locus specific oligonucleotides: a left oligo with a 5′ universal amplification tail, a 5′ phosphorylated middle oligo, and a 5′ phosphorylated right oligo with a 3′ universal amplification tail. To assess fetal fraction, assays were designed against a set of 192 SNP-containing loci on chr1-12, where two middle oligos, differing by one base, were used to query each SNP. SNPs were optimized for minor allele frequency in the HapMa...
PUM
| Property | Measurement | Unit |
|---|---|---|
| nucleic acid proportion | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| frequency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com