Intrinsic chromosomal linkage and disease prediction
a technology of chromosomal linkage and disease prediction, applied in relational databases, database models, instruments, etc., can solve the problems of disassembly of dna variant linkages to human diseases, particularly, and achieve the effect of facilitating the classification of disease risk by genetic markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
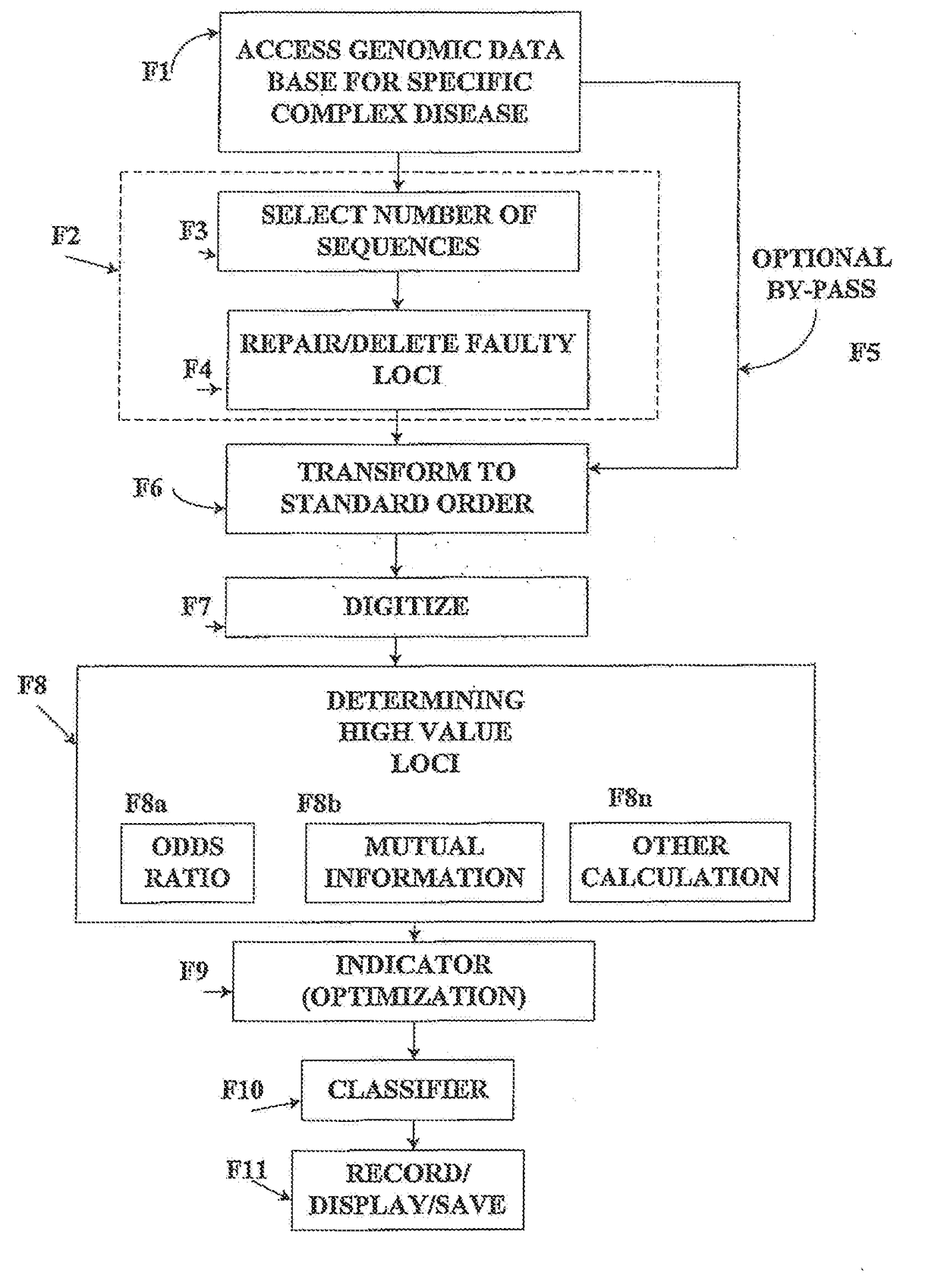
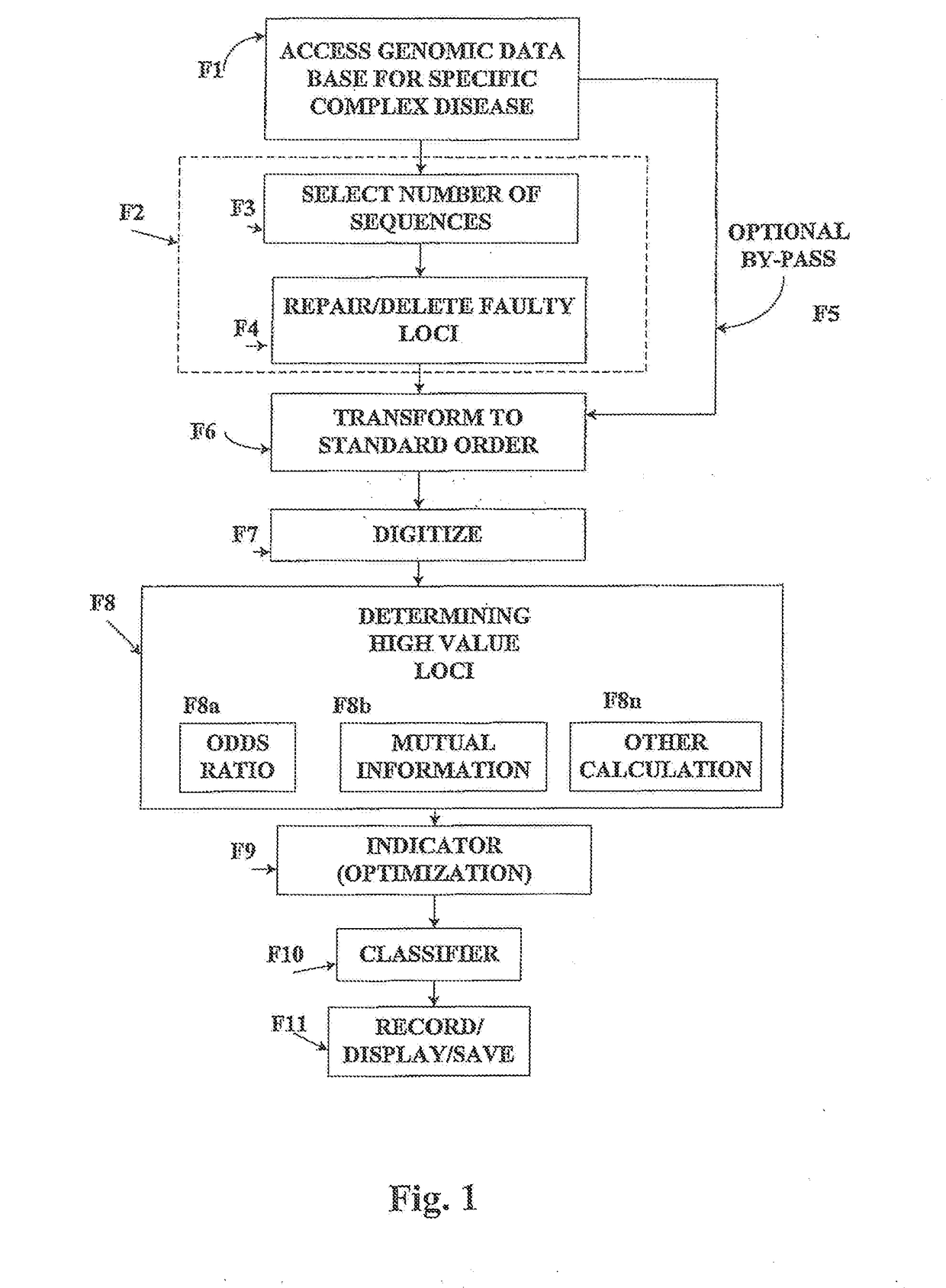
[0035]FIG. 1 depicts the flow diagram of the invention which illustrates the general treatment of a disease / control database. This is illustrated below with the FUSION database.
Technical Details
[0036]An outline of the algorithmic processes is furnished in the flow diagram which describes the components and the steps in the overall algorithmic procedures. Further details of the methods and their algorithmic connections are presented below in relation to FIG. 1.
1. Step F1—Accessing FUSION Database
[0037]In Step F1 of FIG. 1 a genomic database for a complex disease is accessed, which is illustrated by type 2 diabetes (T2D), obtained from the National Institutes of Health.
[0038]The methods of this application are based on clinically linked genomic data. The framework of the methods will be illustrated for type 2 diabetes, T2D: Finland-United States Investigation of NIDDM Genetics (FUSION) study, NIH-dbGap. Details of the database are as follows:
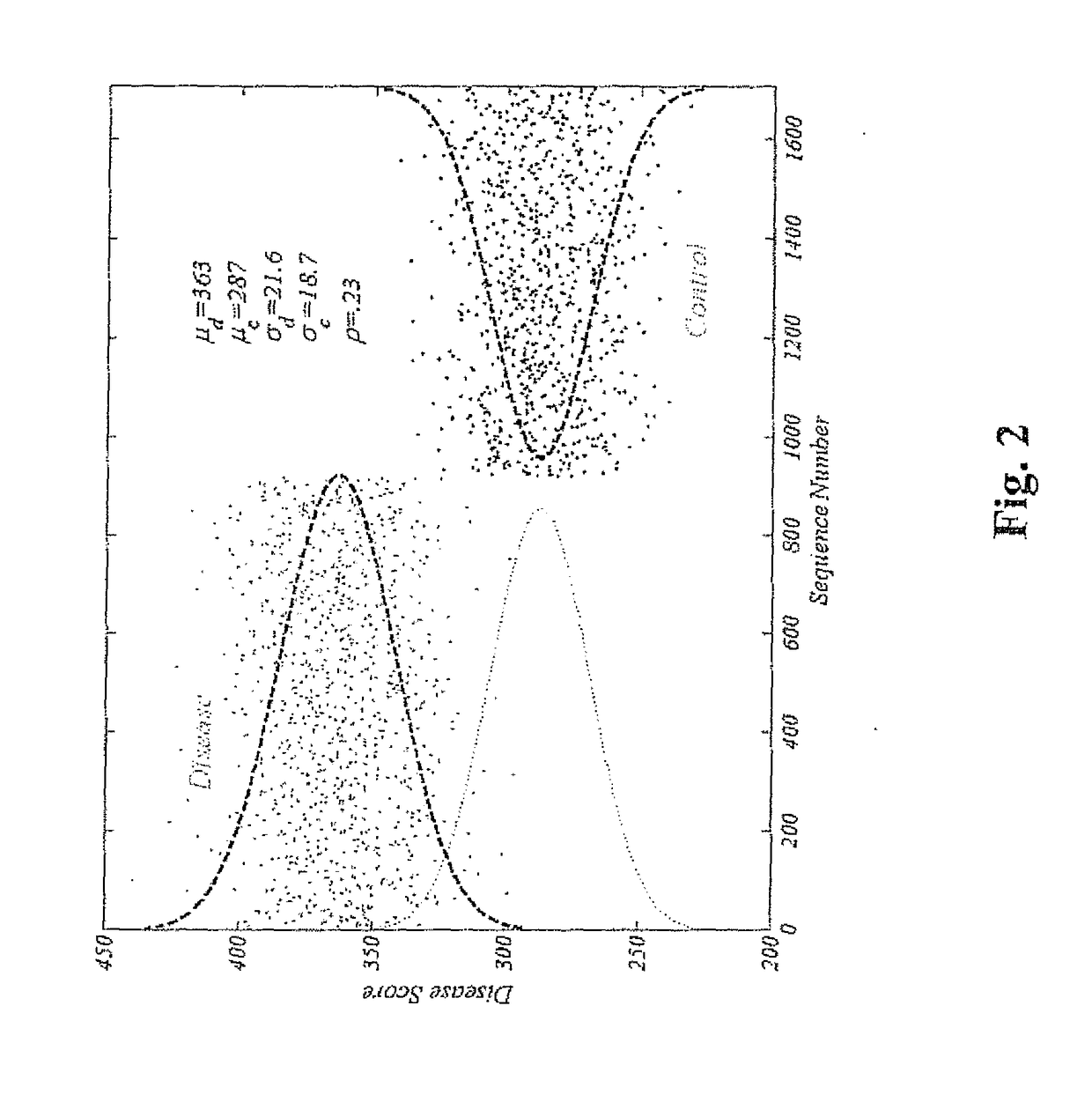
[0039]919 T2D cases, 787 normal glucose tol...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com