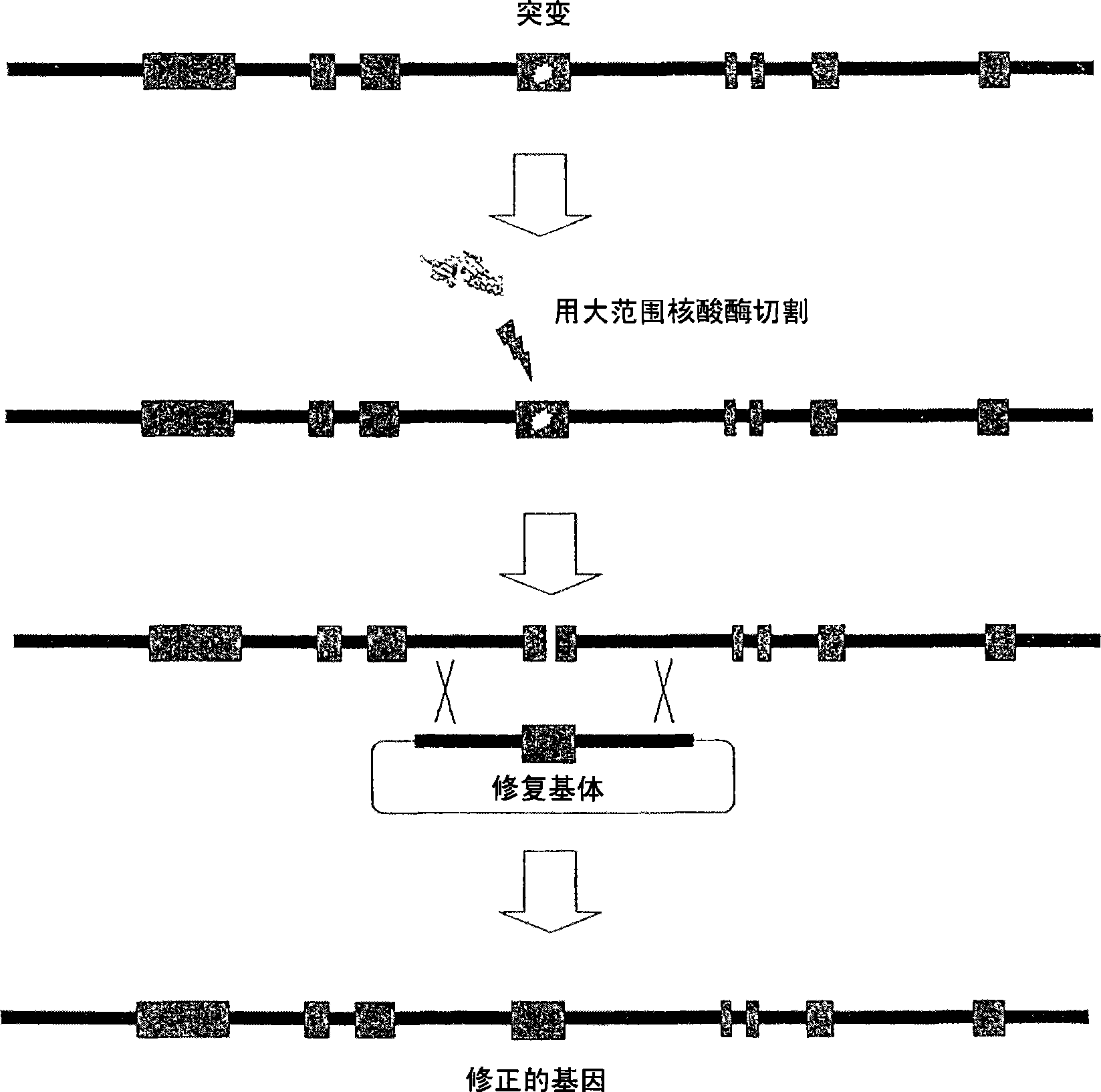
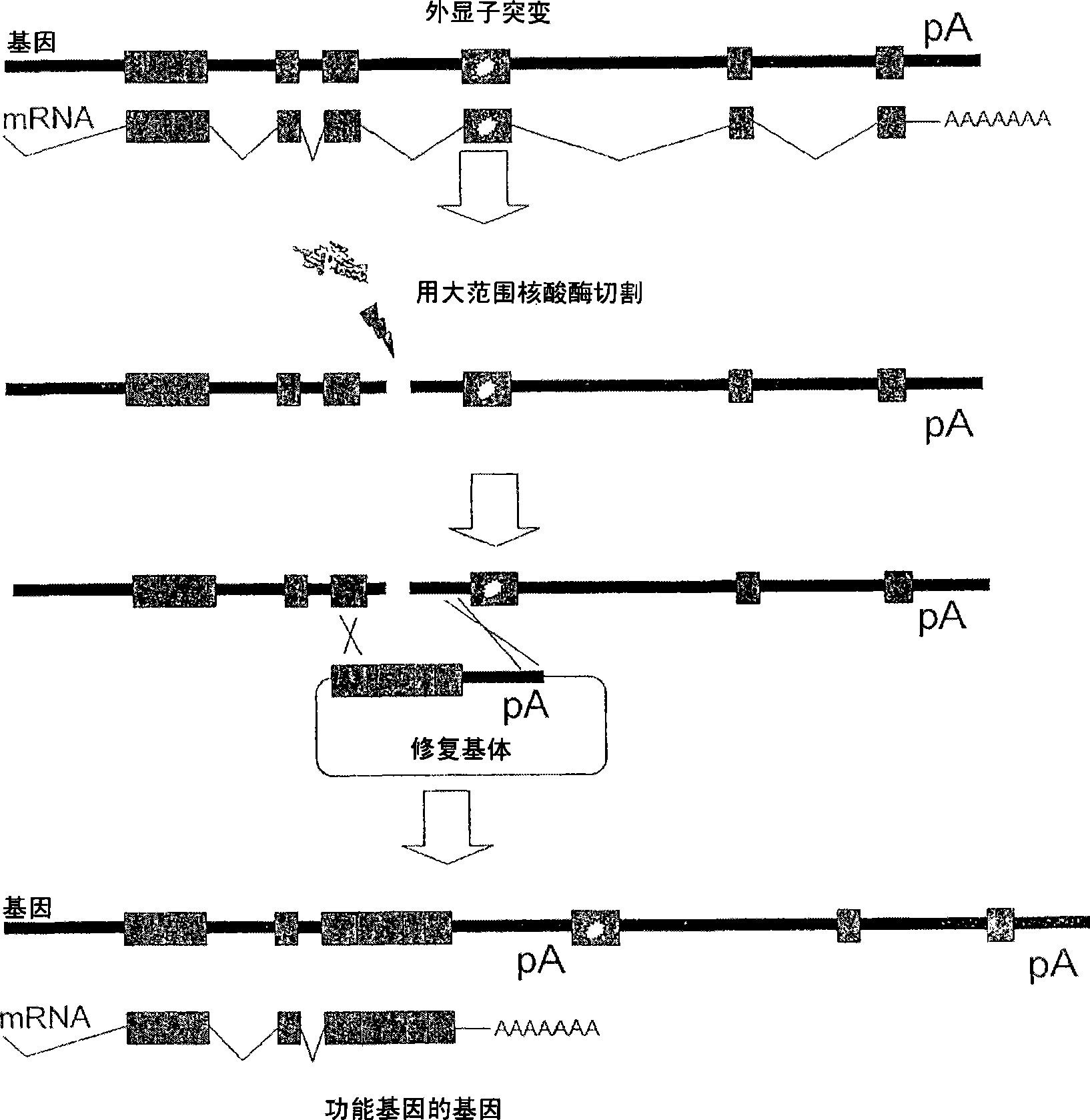
Meganuclease variants cleaving a DNA target sequence from a xp gene and uses thereof
A coloring and variant technology, applied in the direction of stable introduction of foreign DNA into chromosomes, recombinant DNA technology, applications, etc., can solve problems such as difficulty in fully evaluating the sequence range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0200] Example 1 : Transformation of I-CreI variants with specific changes at positions ±8 to ±10
[0201] Methods for generating meganuclease variants, as well as assays based on recombination-induced cleavage in mammalian or yeast cells, were used to screen for variants with altered specificity, which have been described in the literature (PCT Application WO2004 / 067736; Arnould et al., J.Mol.Biol., 2006, 355, 443-458.Epinat et al., N.A.R., 2003, 31, 2952-2962 and Chames et al., Nucleic Acids Res., 2005, 33, e178). These assays yield a functional lacZ reporter that can be monitored by standard methods.
[0202] A) Materials and methods
[0203] a) Construction of libraries Ulib4, Ulib5 and Lib4
[0204] The I-Cre Iwt and I-CreI D75N open reading frames were synthesized as previously described (Epinat et al., 2003, 31, 2952-2962). Mutation D75N was introduced by replacing codon 75 with aac. Three combinatorial libraries (Ulib4, Ulib5, and Lib4) were derived by exchang...
Embodiment 2
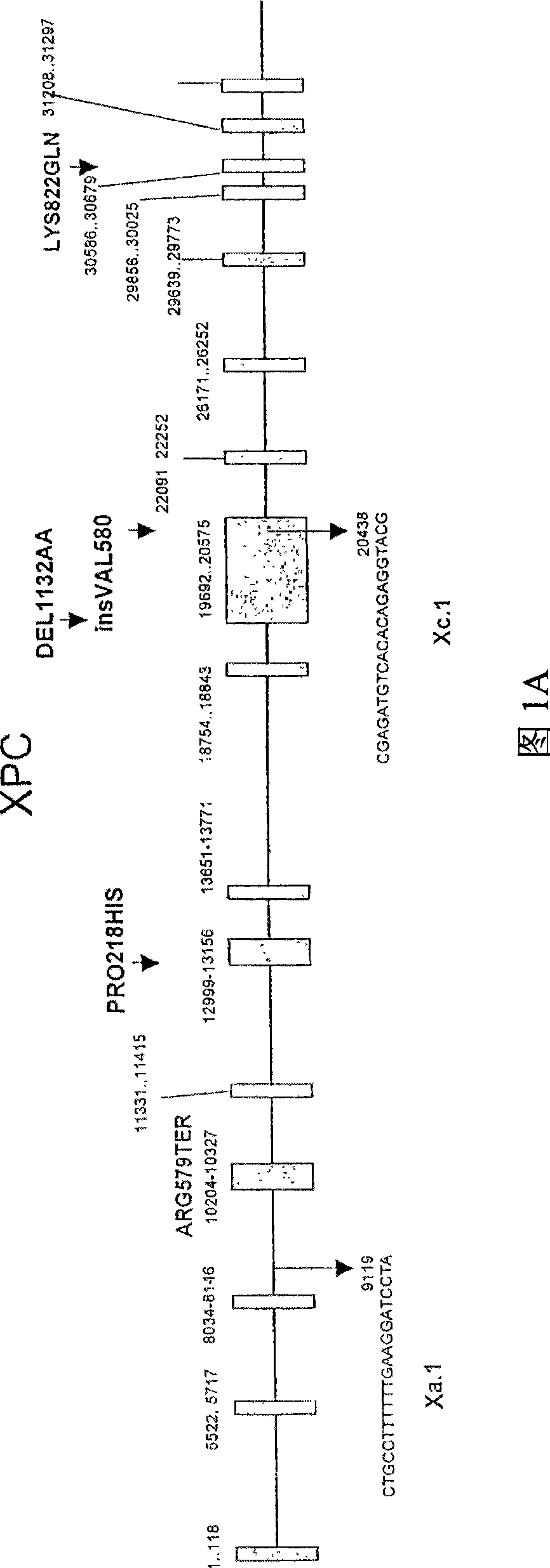
[0232] Example 2 : A strategy for the construction of novel meganucleases that cleave targets from the XPC gene
[0233] The first series of I-CreI variants having at least one substitution at positions 44, 68 and / or 70 of I-CreI and capable of cleaving mutant I-CreI sites with changes in positions ±3 to 5 identified (Arnould et al., J. Mol. Biol., 2006, 355, 443-458). The cleavage patterns of the variants are shown in Figure 7.
[0234] According to the method described in Example 1, it is identified that there is at least one substitution at positions 28, 30, 33 or 28, 33, 38 and 40 of I-CreI, and it is possible to cleave cells with changes in positions ±8 to 10 A second series of I-Crel variants at the mutant I-Crel site. The cleavage patterns of the variants are shown in Figure 6.
[0235] On the same DNA-binding fold, positions 28, 30, 33, 38, and 40 are on one side, and 44, 68, and 70 are on the other, and there is no structural evidence that they work independently...
Embodiment 3
[0242] Example 3 : Preparation of a meganuclease that cleaves Xx.3
[0243] This example shows that the I-Crel mutant can palindromically cleave the Xx.3 DNA target sequence derived from the left portion of the Xx.2 target ( Figure 9 and 23). The target sequence described in this example is a 22bp palindromic sequence. Therefore, only their first 11 nucleotides are described, followed by the suffix _P; for example, target Xa.3 would be annotated as ctgccttttgt_P.
[0244] Xa.3 is similar to 5TTT_P positions ±1, ±2, ±3, ±4, ±5 and ±11, and to 10TGC_P positions ±1, ±2, ±8, ±9, ±10 and ±11.
[0245] Xb.3 is similar to 5GGG_P bits ±1, ±2, ±3, ±4, ±5, and ±11, and to 10GGG_P bits ±1, ±2, ±8, ±9, ±10, and ±11.
[0246] Xc.3 is similar to C1221 bits ±1, ±2, ±3, ±4, ±5, ±7, and ±11, and to 10GAG_P bits ±1, ±2, ±7, ±8, ±9, ± 10 and ±11 bits.
[0247] It is known that wild-type I-CreI allows nucleotide substitutions at positions ±11, ±7 and ±6 (Chevalie et al., J. Mol. Biol., 20...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com