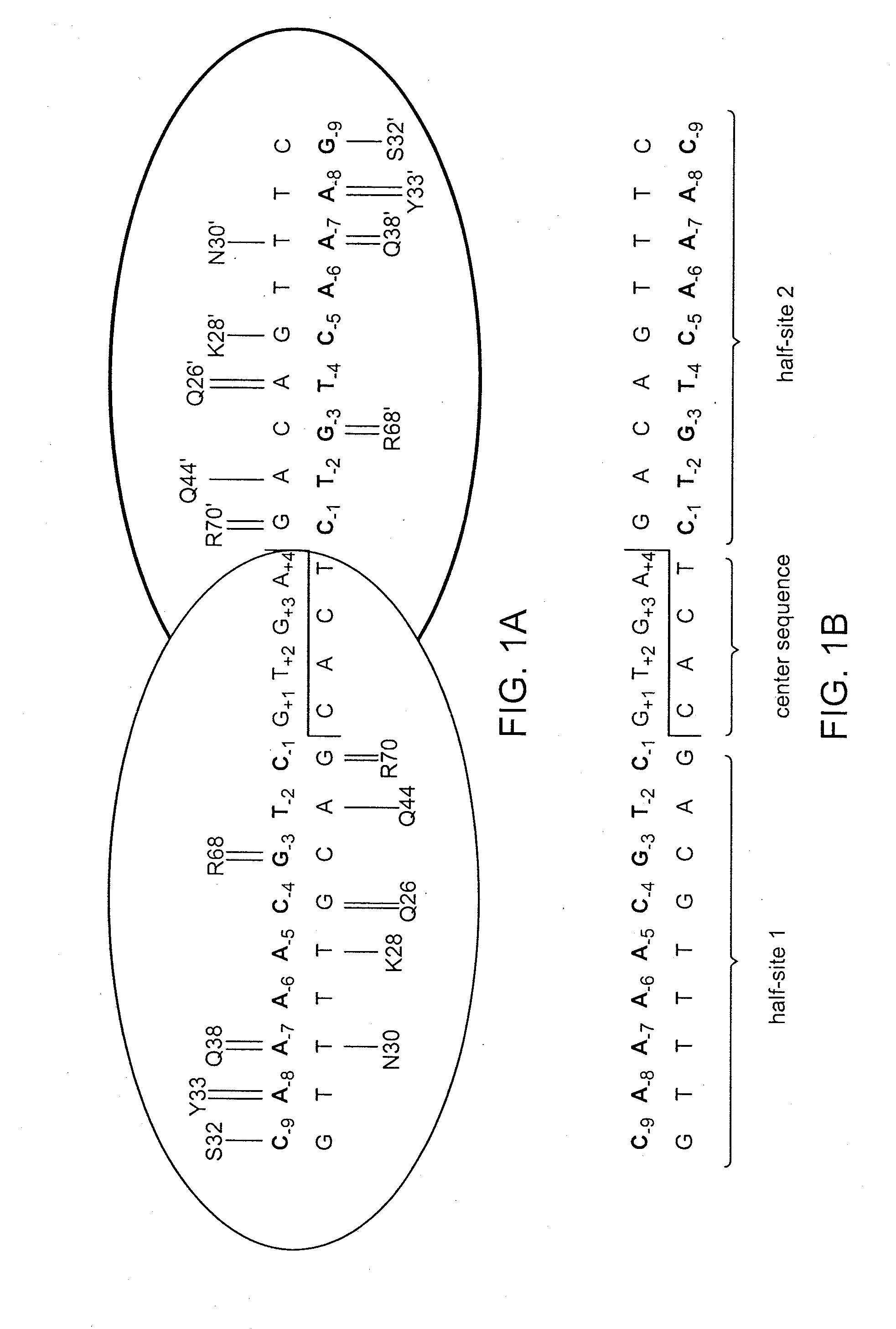
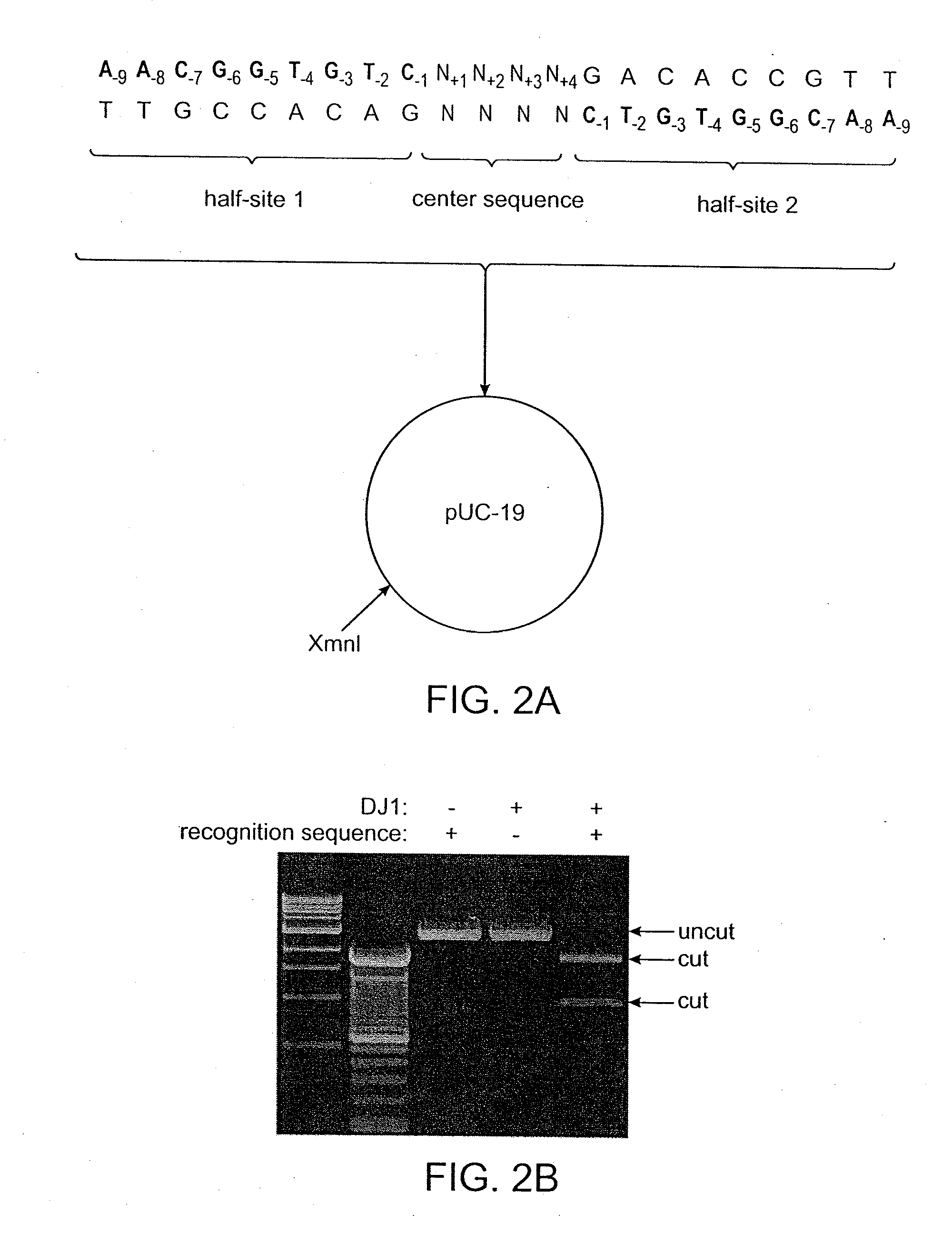
Recognition sequences for i-crei-derived meganucleases and uses thereof
a meganuclease and recognition sequence technology, applied in the field of molecular biology and recombinant nucleic acid technology, can solve the problems of residual non-specific cleavage activity, high mutagenic and toxic, and inability to target gene modifications to unique sites within a chromosomal background, and achieve the effect of more efficient cleavag
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cleavage of an Optimized Meganuclease Recognition Site by a Rationally-Designed, I-CreI-Derived Meganuclease Homodimer in vivo
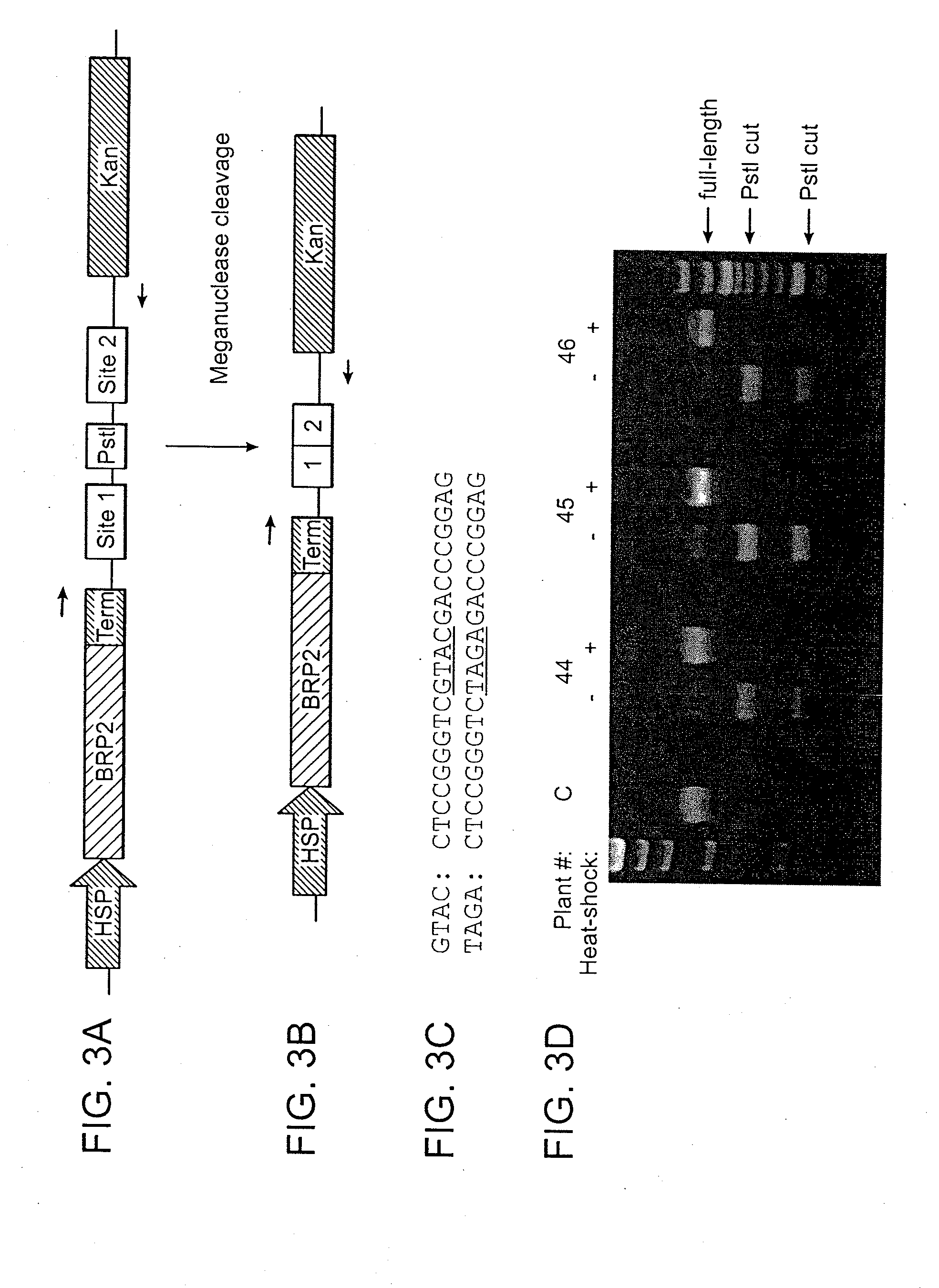
[0106]An engineered meganuclease called BRP2 (SEQ ID NO: 8) was produced using the method disclosed in WO 2007 / 047859. This meganuclease is derived from I-CreI and was engineered to recognize DNA sites that are not recognized by wild-type I-CreI (e.g., BRP2 recognition sequences include SEQ ID NO: 9 and SEQ ID NO: 10, or SEQ ID NO: 11 and SEQ ID NO: 12). To facilitate nuclear localization of the engineered meganuclease, an SV40 nuclear localization signal (NLS, SEQ ID NO: 13) was added to the N-terminus of the protein. Conventional Agrobacterium-mediated transformation procedures were used to transform Arabidopsis thaliana with a T-DNA containing a codon-optimized BRP2 coding sequence (SEQ ID NO: 14). Expression of BRP2 meganuclease was under the control of a Hsp70 promoter and a NOS terminator. A pair of BRP2 recognition sequences were housed on the same T-D...
example 2
Cleavage of an Optimized Meganuclease Recognition Site by a Rationally-Designed, I-CreI-Derived Meganuclease Single-Chain Heterodimer in vivo
[0108]The engineered meganuclease BRP12-SC (SEQ ID NO: 15) was produced in accordance with WO 2007 / 047859, except that this meganuclease is a single-chain heterodimer. As discussed in WO 2007 / 047859, wild-type I-CreI binds to and cleaves DNA as a homodimer. As a consequence, the natural recognition sequence for I-CreI is pseudo-palindromic. The BRP12-SC recognition sequences, however, are non-palindromic (e.g., SEQ ID NO: 16 and SEQ ID NO: 17, or SEQ ID NO: 18 and SEQ ID NO: 19). This necessitates the use of an engineered meganuclease heterodimer comprising a pair of subunits each of which recognizes one half-site within the full-length recognition sequence. In the case of BRP12-SC, the two engineered meganuclease monomers are physically linked to one another using an amino acid linker to produce a single-chain heterodimer. This linker comprise...
example 3
Cleavage of an Optimized Meganuclease Recognition Site by a Rationally-Designed, I-CreI-Derived Meganuclease Homodimer In Vitro
[0111]The BRP2 meganuclease described in Example 1 (SEQ ID NO: 8) was expressed in E. coli and purified as in Example 1 of WO 2007 / 047859. The purified meganuclease was then added at varying concentrations to reactions containing plasmids harboring BRP2 recognition sequences with either a GTAC or TAGA center sequence (0.25 picomoles of plasmid substrate in 25 microliters of SA buffer: 25 mM Tris-HCL, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 5 mM EDTA). Reactions were incubated at 37° C. for 2 hours and were then visualized by gel electrophoresis and the percentage of each plasmid substrate cleaved by the meganuclease was plotted as a function of meganuclease concentration (FIG. 5). It was found that the plasmid substrate with the TAGA center sequence was cleaved by the meganuclease in vitro, but that cleavage of this substrate required a far higher concentration of ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com