Method for identification and monitoring of epigenetic modifications
A biological sample and imprinting technology, applied in the field of identification and monitoring of epigenetic modifications, can solve the problems of little progress and limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1: ChIP Chip for Discovery and Monitoring of Imprinted Genes TM
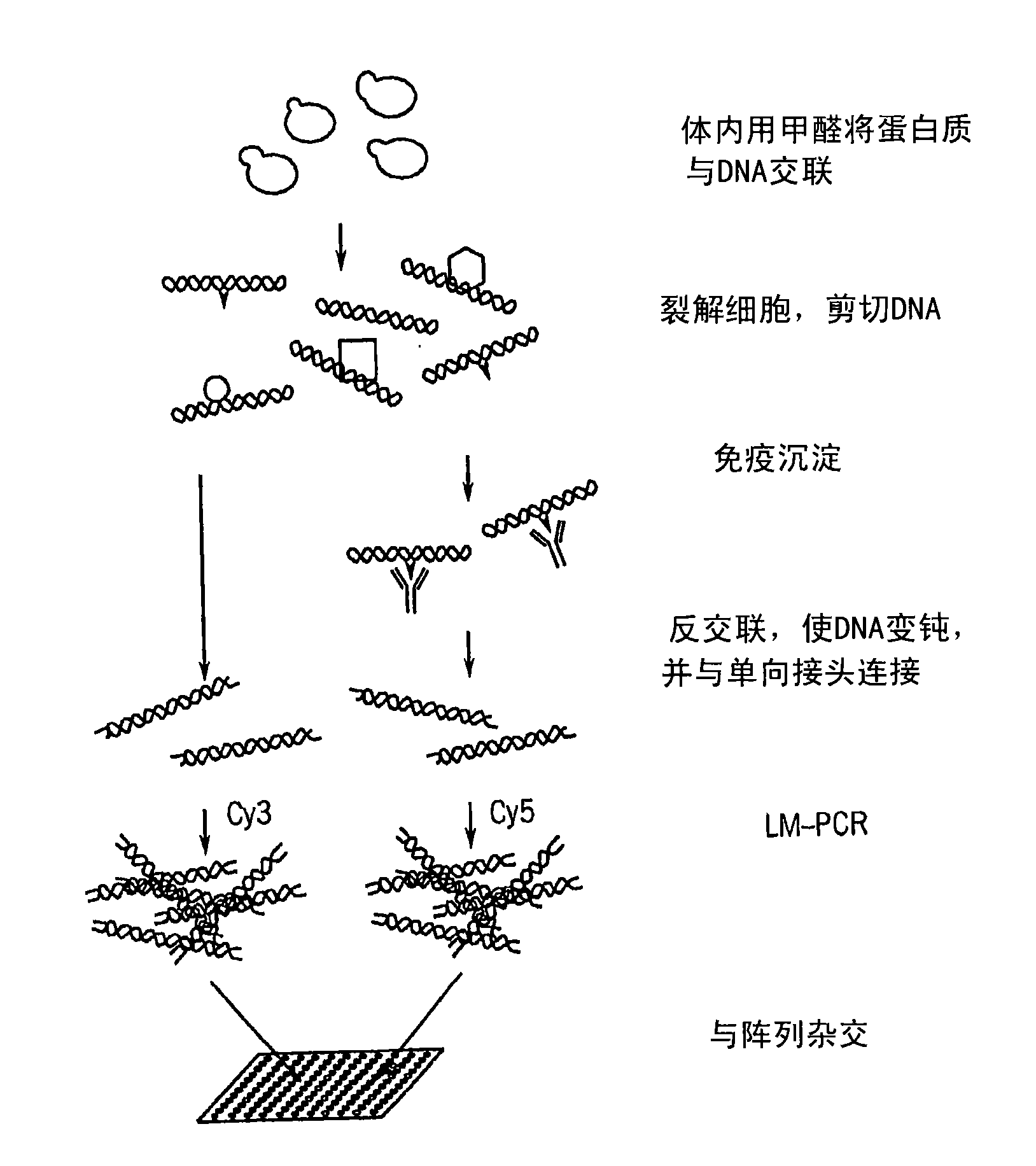
[0068] To determine genomic regions containing imprinted genes, applicants performed a ChIP Chip TM Experimentally, regions with overlapping open and closed chromatin marks were identified by their association with preferred modified histone marks, such as AcK9H3, 3MeK9H3, 2MeK9H3 and RNA polymerase II.
[0069] In short, to perform ChIP Chip TM In the experiment, cells were chemically treated to cross-link DNA-binding proteins at their binding sites. DNA was isolated from these cells, fragmented, and enriched by immunoprecipitation with antibodies against modified histones and the RNA polymerase of interest (see generally Ng et al., Genes Dev. 16, 806-819 (2002); Wyrick et al., Science 290, 2306-2309 (2002); Weinmann et al., Genes Dev. 16, 235-244 (2002)).
[0070] These enriched DNAs were then amplified by LM-PCR. Such as figure 1 As shown, the methods of the present invention are not limi...
Embodiment 2
[0073] Example 2: Microarray Analysis of Coexisting Silent and Active Chromatin Marks as a High-Throughput Method to Identify Newly Imprinted Genes
[0074] Regardless of the CpG island methylation status in a particular cell type, Applicants performed a series of microarray experiments in the presence or absence of CpG island or GC-rich sequences to efficiently screen or identify concurrent "active" or " Silent" chromatin marks.
[0075] Chromatin immunoprecipitation
[0076] Chromatin immunoprecipitation (ChIP) was performed in cultured HeLA and 293 cells using the known ChIP protocol electronically published by Farnham Laboratories, published on December 25, 2005 as follows: http: / / genomecenter.ucdavis.edu / farnham / farnham / protocols / chips.html , a protocol incorporating ligation-mediated PCR (LMPCR) based on the method of Ren et al. ((2000) Science 290, p. 2306) (both articles are incorporated herein by reference in their entirety).
[0077] Materials for ChIP analysis ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com