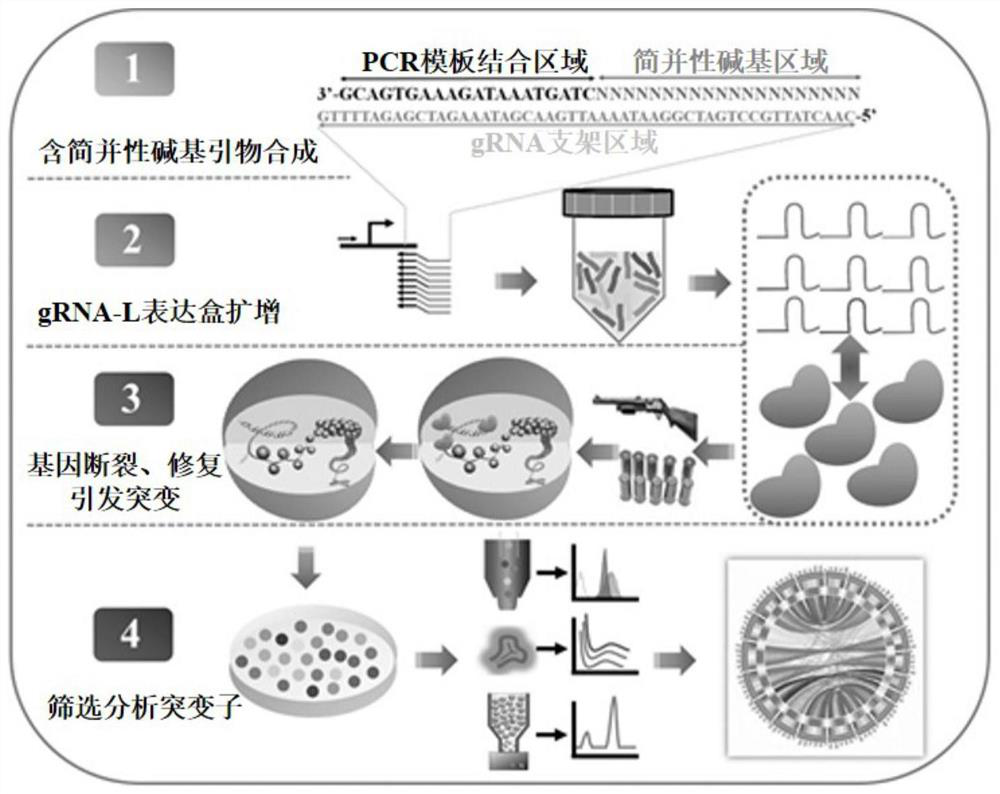
Whole-genome random mutation method based on CRISPR-Cas system and application of whole-genome random mutation method
A whole-genome, random mutation technology, applied in the field of bioengineering, can solve the problems of destroying the fidelity of genome replication, poor stability of mutants, single mutation type, etc., reaching single-cell mutation sites with diverse mutation types and high mutation rate , the effect of simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] 1.1 Construction of SpCas9-NG expression vector pCas9-NG:
[0036] Using the pTCL vector (Addgene: #43802) as a template, use two pairs of primers pCas9-NG-F1 / pCas9-NG-R1 and pCas9-NG-F2 / pCas9-NG-R2 to amplify, respectively, to mutate 5 DNA sequences of SpCas9 The codons, specifically L1111R / D1135V / A1322R / R1335V / T1337R, allow the mutated Cas protein Cas9-NG to recognize the NG sequence and expand the Cas protein target; then, the two PCR fragments are circularized into plasmid pCas9 by Gibson assembly -NG.
[0037] Wherein, the nucleotide sequence of pCas9-NG-F1 is shown in SEQ ID NO.5:
[0038] GCTGATCGCACGCAAAAAAGATTGGGACCCCAAAGAAATACGGCGGATTCGTTTCTCCTACAGTCGCTTAC;
[0039] The nucleotide sequence of pCas9-NG-R1 is shown in SEQ ID NO.6:
[0040] CTTTCTGTCTATGGTGGTGTCGAAGTACTTGAAGGCTCGAGGCGCGCCCAAGTTGGTC;
[0041] The nucleotide sequence of pCas9-NG-F2 is shown in SEQ ID NO.7:
[0042] ACCACCATAGACAGAAAGGTGT ACCGCTCTCACAAAGGAGGTCCTG;
[0043] The nucleotide seque...
Embodiment 2
[0057] 2.1 The method of the present invention is applied to the directed evolution and efficiency verification of β-carotene-producing Saccharomyces cerevisiae
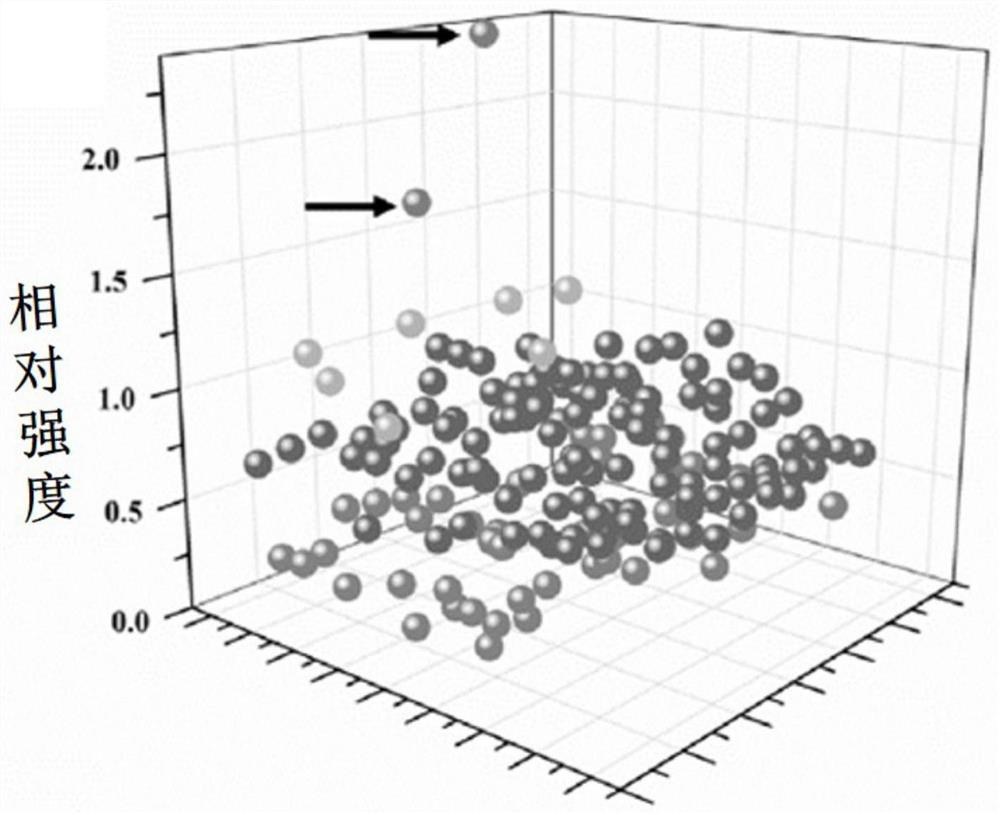
[0058] For a strain of Saccharomyces cerevisiae producing β-carotene, the plasmid pCas9-NG was electrotransformed into the cells to obtain strain C0, and then 1 μg of each PCR-amplified toolbox random library fragment expressing gRNA and pSCM linearized plasmid backbone were electrotransformed into C0 For competent cells, the transformed CO competent cells were serially diluted and spread on SD-Leu-Ura plates. Select a plate with 50-200 growing colonies, and finally select a plate with 176 colonies. Compared with the ability to produce β-carotene from the starting strain C0, the β-carotene production capacity of the final 2 bacterial colonies is significantly improved (such as figure 2 shown).
[0059] 2.2 The method of the present invention is applied to the iterative evolution of Saccharomyces cerevisiae producin...
Embodiment 3
[0063] 3.1 Genome-wide mutation analysis of iterative evolution strains
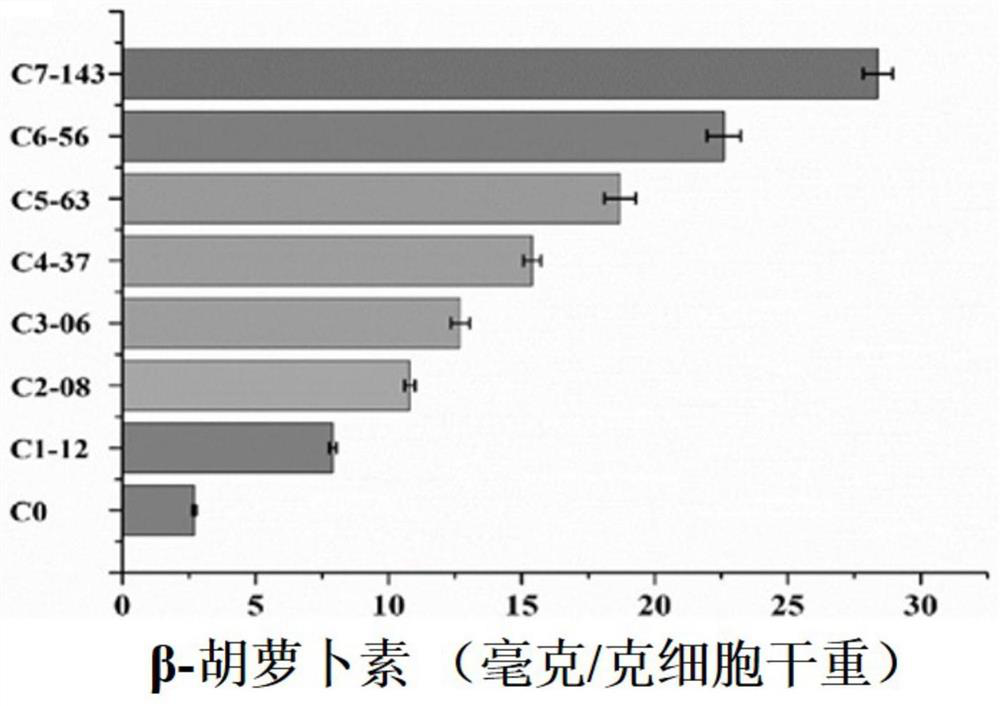
[0064] Using the third-generation genome sequencing platform, analyze the final evolution strain C7-143, process evolution strains C3-06 and C5-63, and the whole genome sequence of the starting strain C0, and compare the mutation sites of the evolution strains compared with C0. The analysis results are as follows Figure 4 and Figure 5 shown.
[0065] On average, each round of evolution can cause 122 mutations in the Saccharomyces cerevisiae genome, and the mutation types are diverse, including base insertion / deletion, duplication, inversion and chromosomal rearrangement.
[0066] Mutation sites are diversified in chromosome locations, distributed in exons, introns, spacers, 3' non-coding regions, and 5' non-coding regions.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com