Transformed CRISPR/SaCas9 (clustered regularly interspaced short palindromic repeat/streptococcus pyogenes Cas9) system targeting at hepatitis B virus and application of system
A virus, enh2-pa1at technology, applied in the modified CRISPR/SaCas9 system targeting hepatitis B virus and its application field, which can solve the problems of single real-time quantitative PCR and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] [Example 1] Design, synthesis and eukaryotic expression vector construction of gRNA sequences targeting 22 different genotypes of HBV conservative sequences
[0060] 1. Selection and design of gRNA targeting HBV DNA
[0061] Using the D-type HBV (Genebank ID: V01460.1) as the reference sequence, the target site with a higher score on the HBV DNA was found through the Crisprdesign tool on crispr.mit.edu, and the target site sequence was in the form of 5'-(20N )-NNGRRT3' or 5'-ARRCNN(20N)-3'. Different from previous designs, the present invention compares the HBV DNA sequences of 22 different genotypes, and selects the gRNA sequences located in highly conserved regions from the found target sites. These conserved regions themselves are also genes in the replication process. Regions with low mutation rates. At the same time, the human genome sequence was compared, gRNAs with high homology to the human genome sequence were excluded, and a series of gRNA sequences and targ...
Embodiment 2
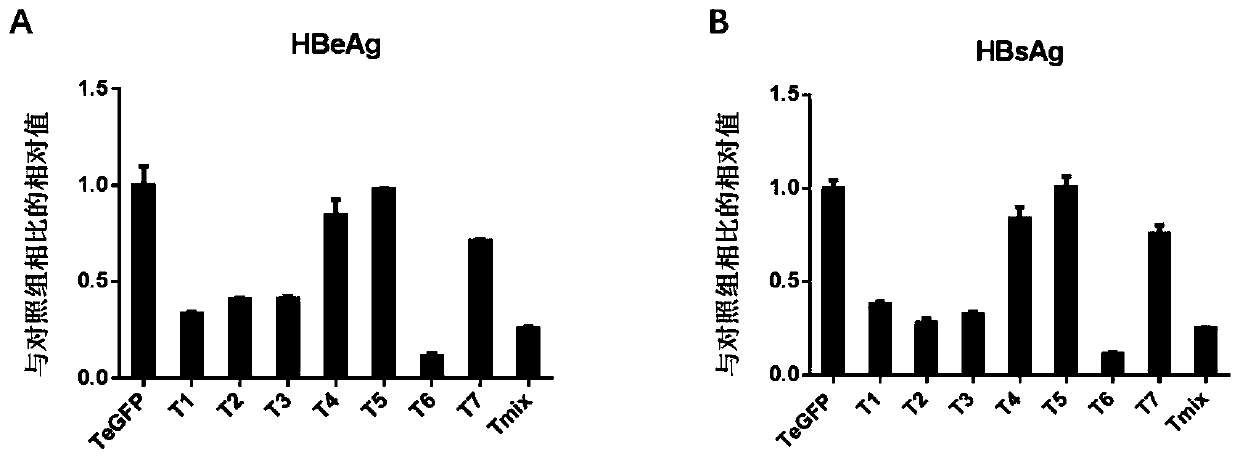
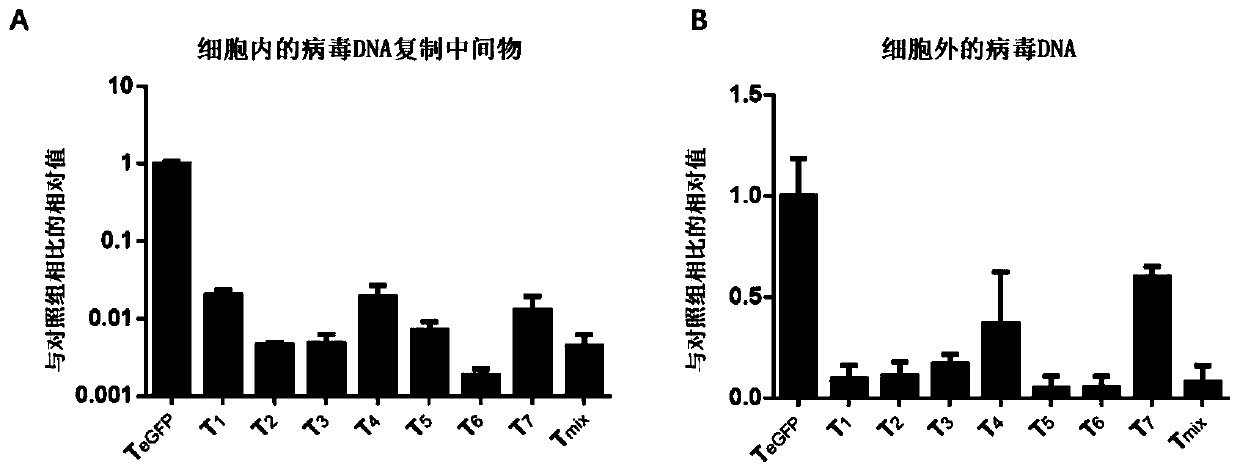
[0078] [Example 2] Verifying the inhibition of HBV replication by the gRNA-guided CRISPR / Cas9 system of the present invention at the cellular level
[0079] In order to verify whether the gRNA-guided CRISPR / SaCas9 system designed in the present invention has the ability to resist HBV replication, the clone of the constructed gRNA will be combined with the replicon prcccDNA (genotype D; Genebank ID: V01460. 1), pCMV-Cre was co-transfected into the liver cancer cell line Huh7, and another gRNA targeting GFP (T GFP ) were used as a control group. To normalize the transfection efficiency between different wells, each well was transfected with the same amount of β-galactosidase expression plasmid pSVβ-gal. After 48 hours of transfection, it was observed that the growth state of the Huh7 cell line was good, and the culture supernatant was taken to measure the expression levels of hepatitis B virus surface antigen (HBsAg) and hepatitis B virus e antigen (HBeAg). The results showed ...
Embodiment 3
[0161] [Example 3] Differential expression levels of different candidate promoters in liver-derived and non-liver-derived cell lines
[0162] 1. According to the genome structure of D-type HBV (Genebank ID: V01460.1), select the corresponding four promoters Enh1 / X, Enh2 / C, preS1, preS2, and one enhancer Enh2 as candidates; according to human and rat genomes Information selected three promoters PPck (Gene ID: 362282), Palb (Gene ID: 111832673), Pa1AT (Gene ID: 5265) with liver-specific expression, and one enhancer Ealb as candidates. Finally, 13 promoter combinations were selected: Enh1 / X, Enh2 / C, preS1, preS2, PPck, Ealb-PPck, Enh2-PPck, Palb, Ealb-Palb, Enh2-Palb, Pa1AT, Ealb-Pa1AT, Enh2-Pa1AT.
[0163] 2. Ligate the above promoter fragment and CMV promoter with the pGL3 luciferase reporter plasmid treated with Sac1 enzyme and Hind3 enzyme;
[0164] 3. The 14 plasmids constructed in the above 2 were co-transfected with the plasmid phRL-TK carrying the Renilla luciferase gene...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com