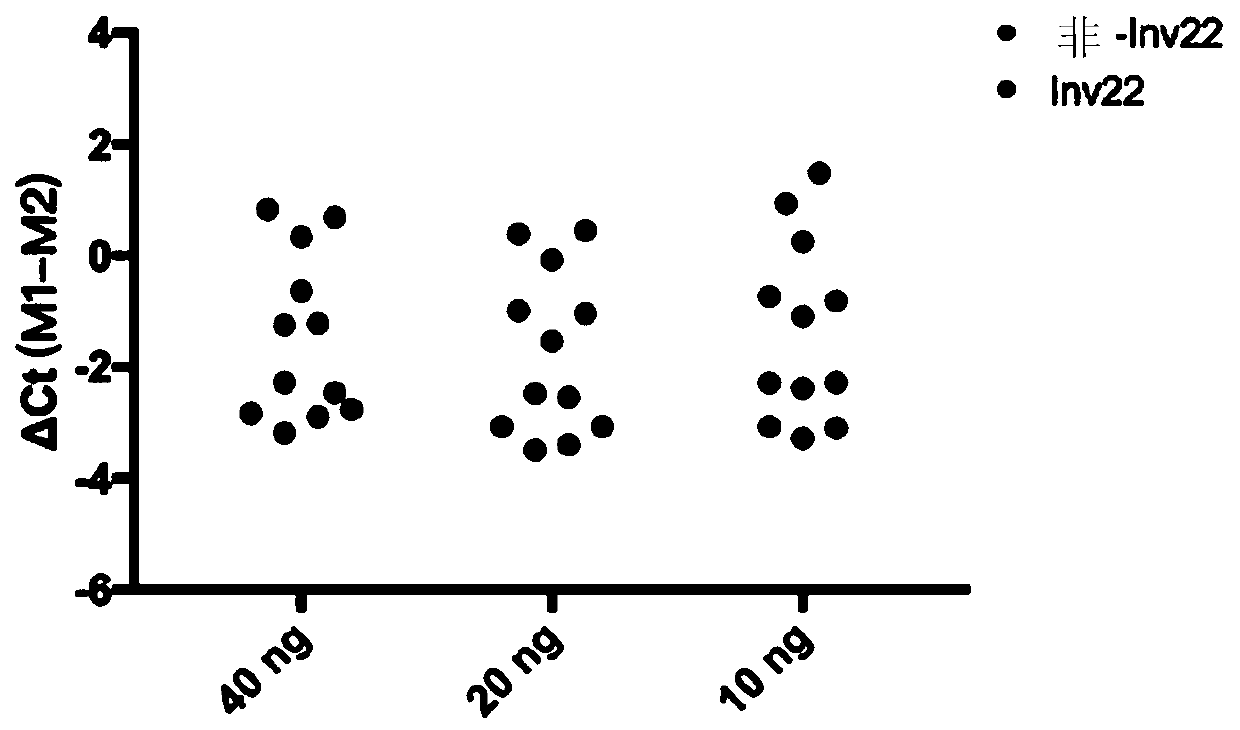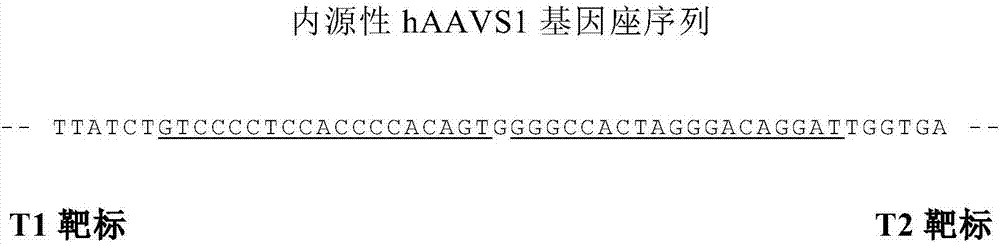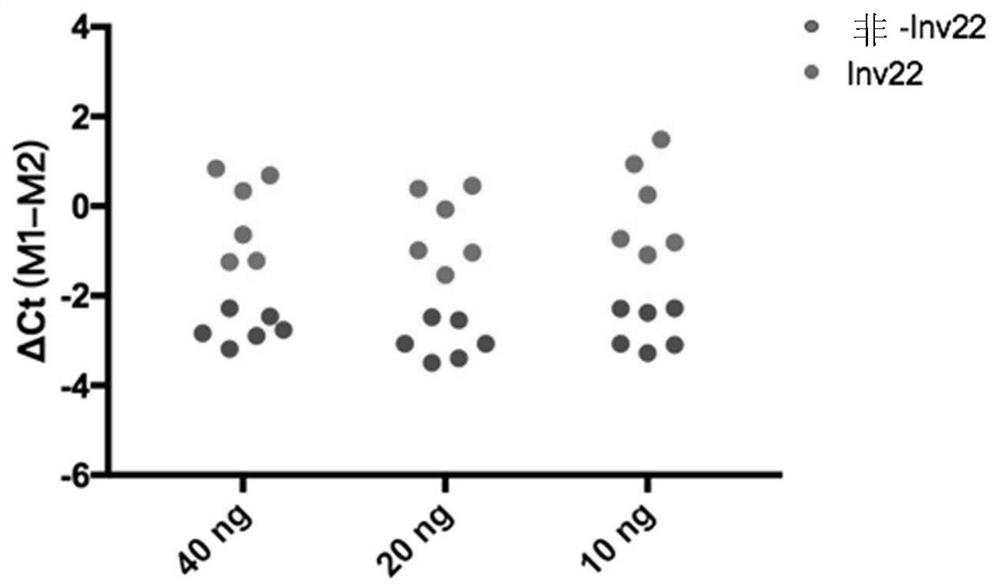Patents
Literature
31 results about "Chromosomal rearrangement" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In genetics, a chromosomal rearrangement is a mutation that is a type of chromosome abnormality involving a change in the structure of the native chromosome. Such changes may involve several different classes of events, like deletions, duplications, inversions, and translocations. Usually, these events are caused by a breakage in the DNA double helices at two different locations, followed by a rejoining of the broken ends to produce a new chromosomal arrangement of genes, different from the gene order of the chromosomes before they were broken. Structural chromosomal abnormalities are estimated to occur in around 0.5% of newborn infants.
High resolution chromosomal mapping
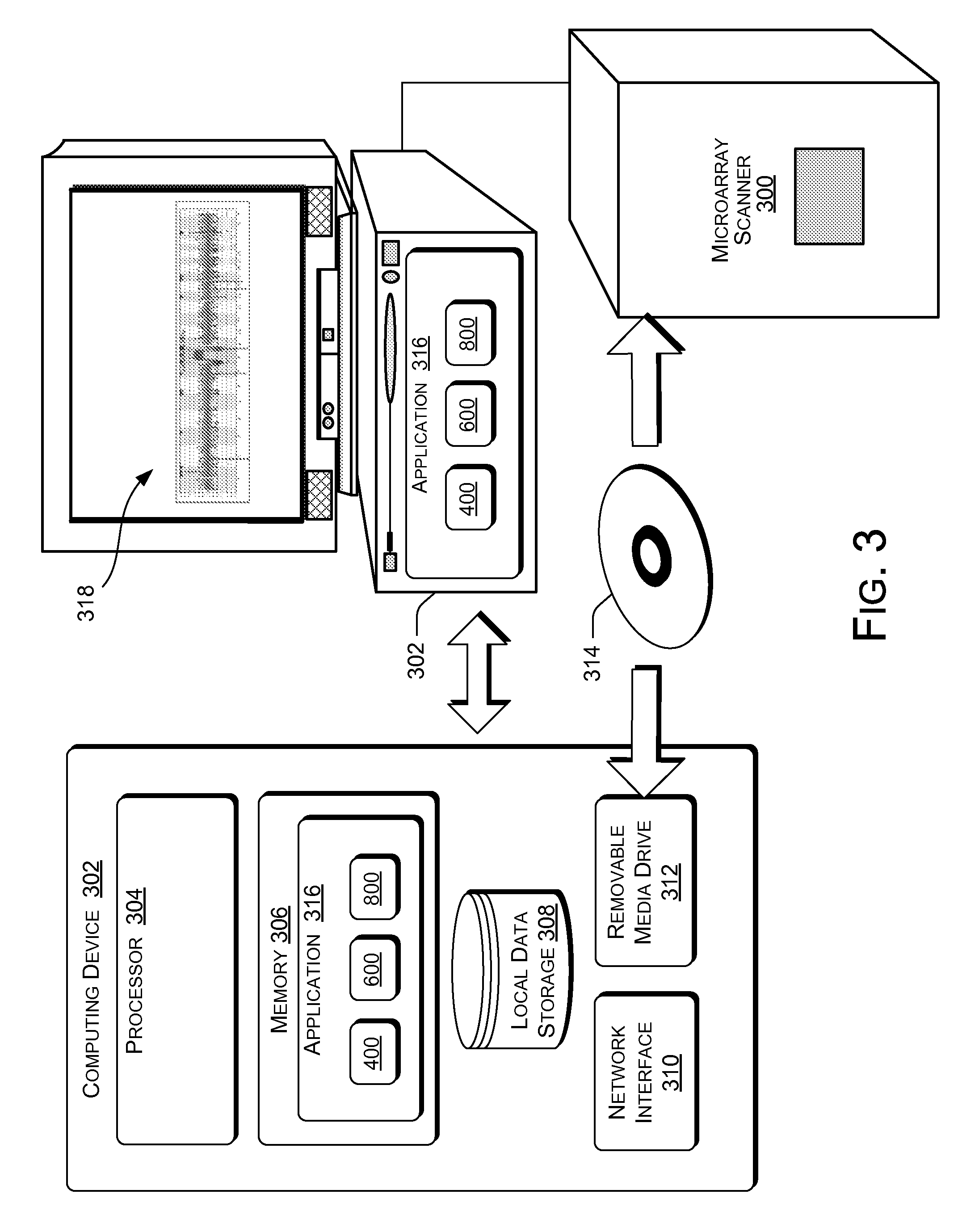
ActiveUS20070238105A1Bioreactor/fermenter combinationsBiological substance pretreatmentsStructural analysisGenome
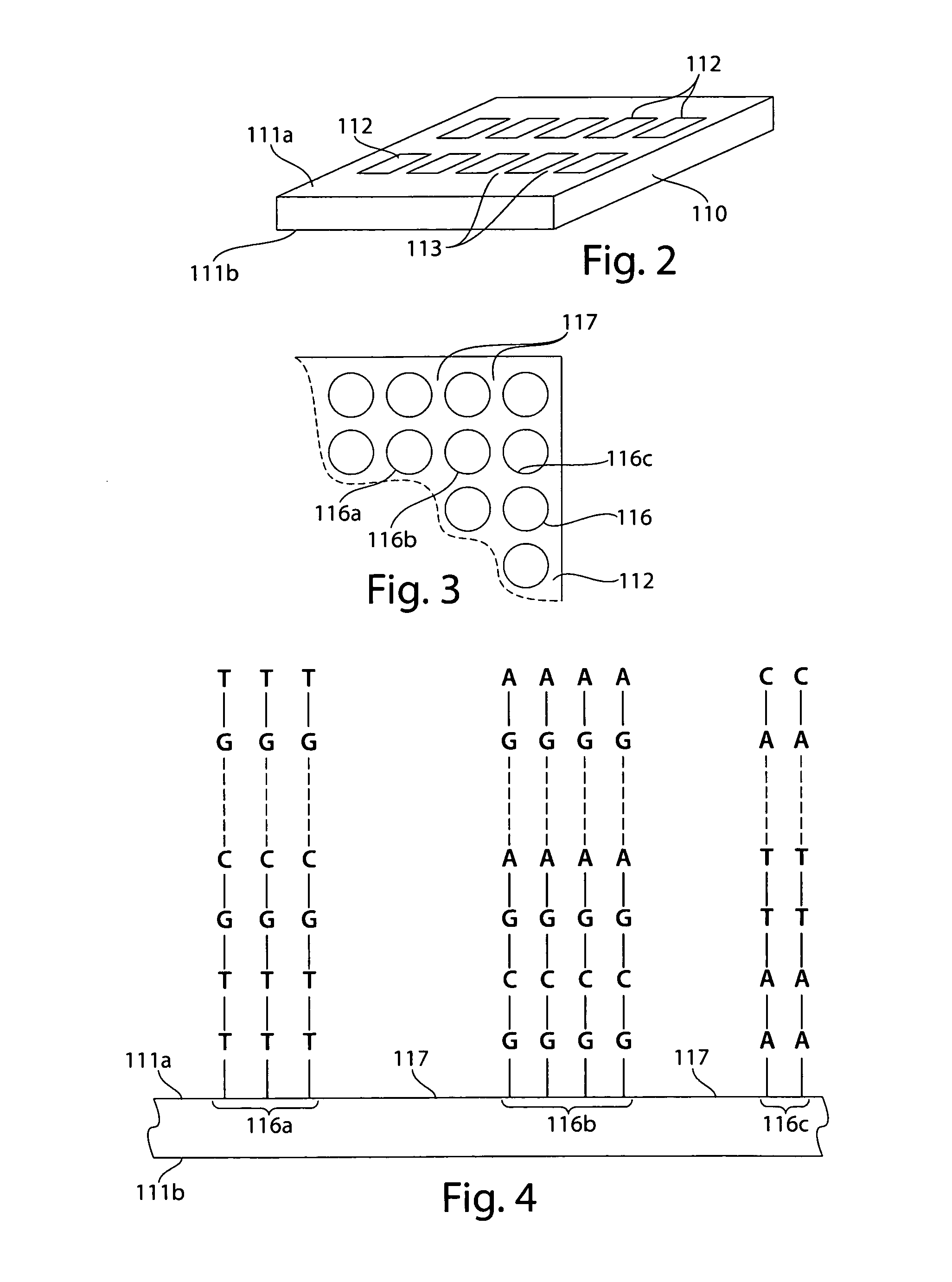
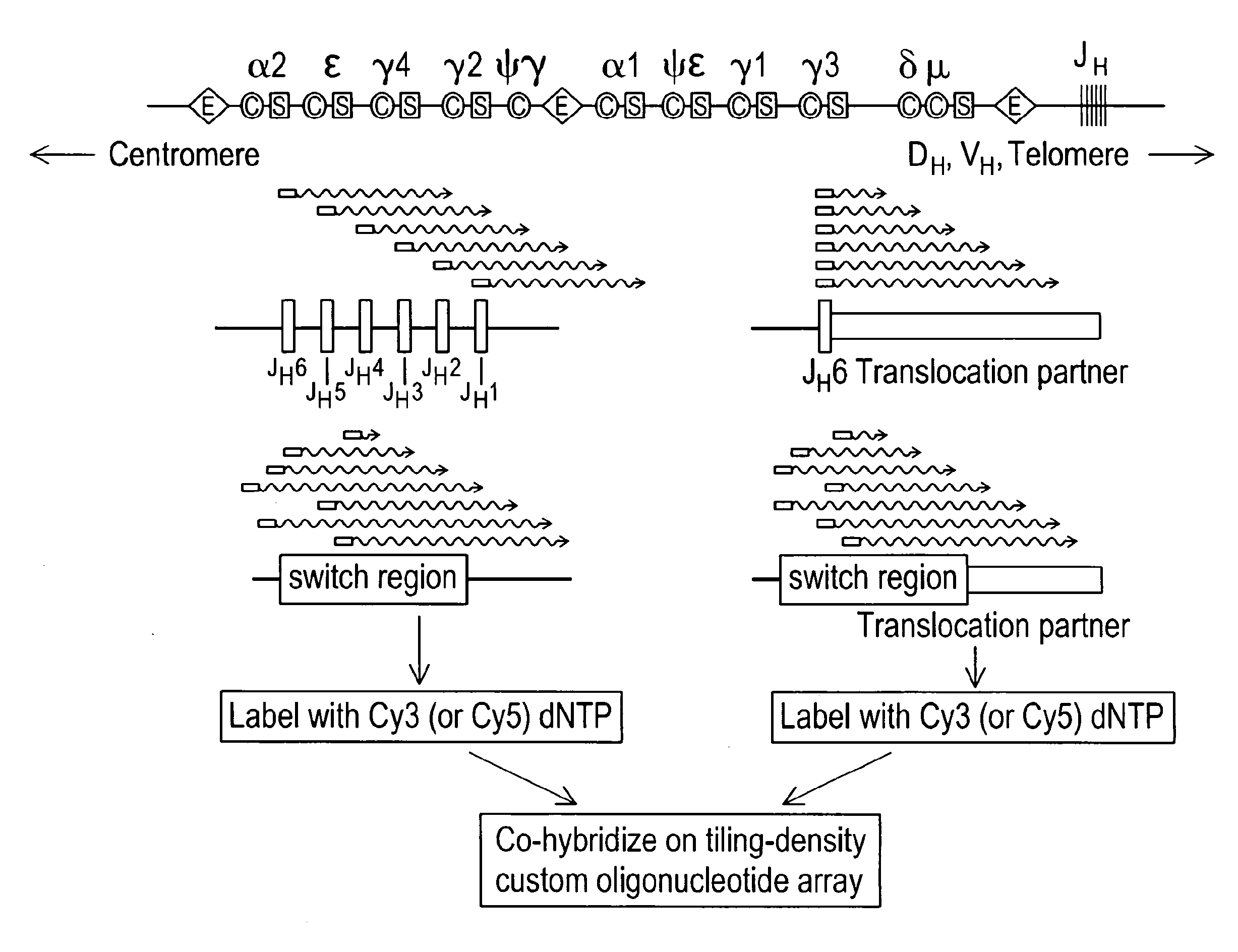
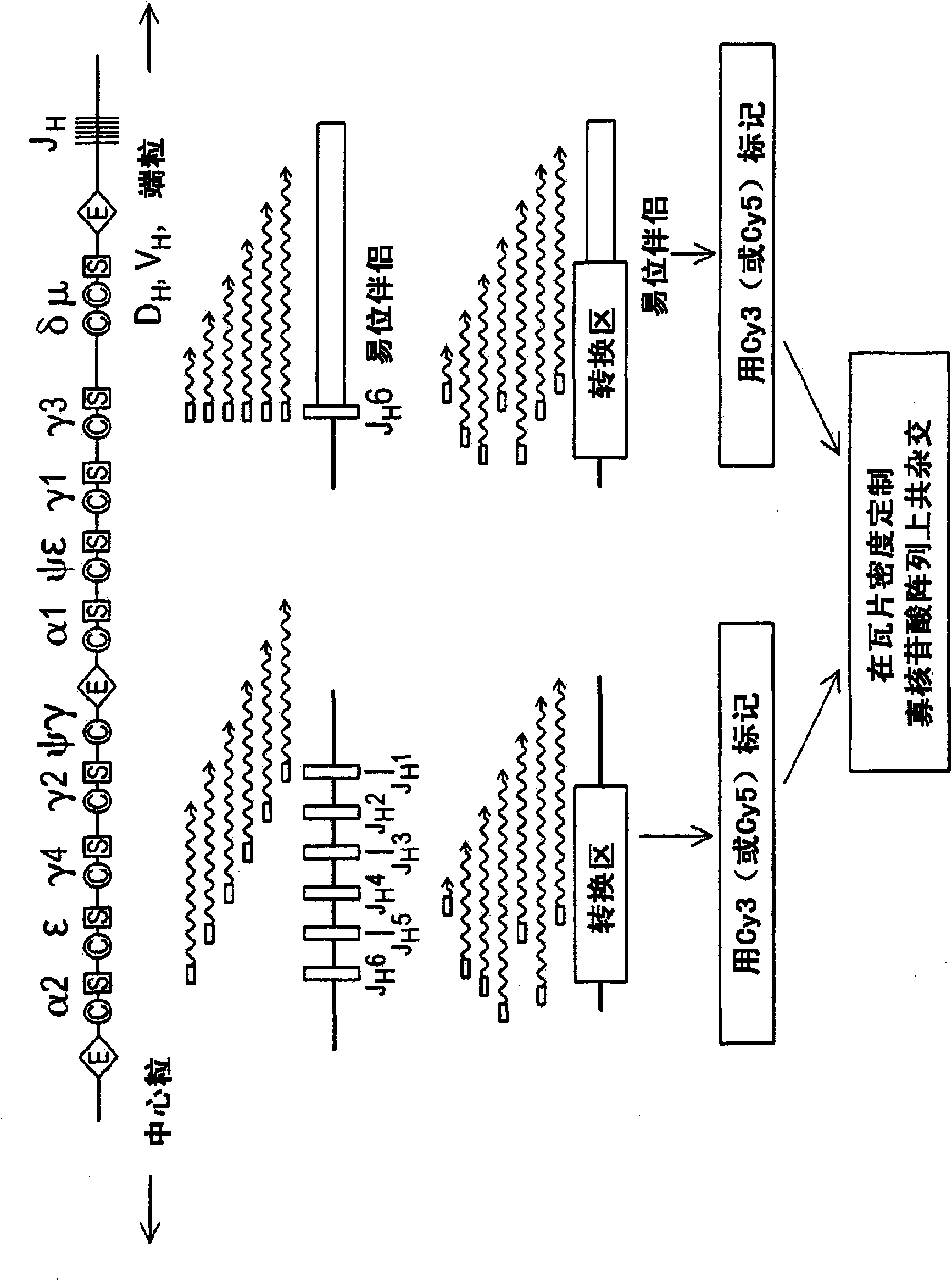
The present invention generally relates to spatial and structural genomic analysis compositions, which can be used for the mapping of chromosomes and structural analyses of chromosomal rearrangements, including the entire chromosome, as well as specific portions or regions of interest of the chromosomes. In some embodiments, multiple portions of the genome can be distinguished, for instance, using a first detection entity and a second detection entity different from the first detection entity. The detection entities may be immobilized relative to oligonucleotides, which may be selected to bind to different locations within the chromosome. For instance, the oligonucleotides may be at least substantially complementary to the chromosome, e.g., substantially complementary to a specific location of the chromosome.
Owner:AGILENT TECH INC
High resolution chromosomal mapping
ActiveUS8058055B2Bioreactor/fermenter combinationsBiological substance pretreatmentsStructural analysisGenome
Owner:AGILENT TECH INC
Methods and Compositions for the Prediction of Response to Trastuzumab Containing Chemotherapy Regimen in Malignant Neoplasia
InactiveUS20090280493A1Large capacityEasy to explainMicrobiological testing/measurementMaterial analysisDiseaseNeoplastic tissue
The invention relates to methods and compositions for the prediction, diagnosis, prognosis, prevention and treatment of neoplastic disease. Neoplastic disease is often caused by chromosomal rearrangements which lead to over- or underexpression of the rearranged genes. The invention discloses genes which are overexpressed in neoplastic tissue and are useful as diagnostic markers and targets for treatment. Methods are disclosed for predicting, diagnosing and prognosing as well as preventing and treating neoplastic disease.
Owner:SIEMENS HEALTHCARE DIAGNOSTICS INC
Identification and treatment of estrogen responsive prostate tumors
InactiveUS20090036415A1Great chance of notDecrease in tumor massBiocideOrganic active ingredientsAgonistEstrogen Receptor Antagonists
The present invention is directed to specific chromosomal rearrangements that are associated with prostate tumors that respond to compounds acting at estrogen receptors. Patients having the TMPRSS2-ERG fusion, may be treated with agonists of the estrogen beta receptor or antagonists of the estrogen alpha receptor.
Owner:THE BRIGHAM & WOMEN S HOSPITAL INC
Microrna-based methods and compositions for the diagnosis, prognosis and treatment of tumor involving chromosomal rearrangements
Owner:UNIV OF BREMEN
Quantitative multiplex amplification on a genomic scale, and applications for detecting genomic rearrangements
InactiveUS20050244830A1Sugar derivativesMicrobiological testing/measurementHuman DNA sequencingMultiplex
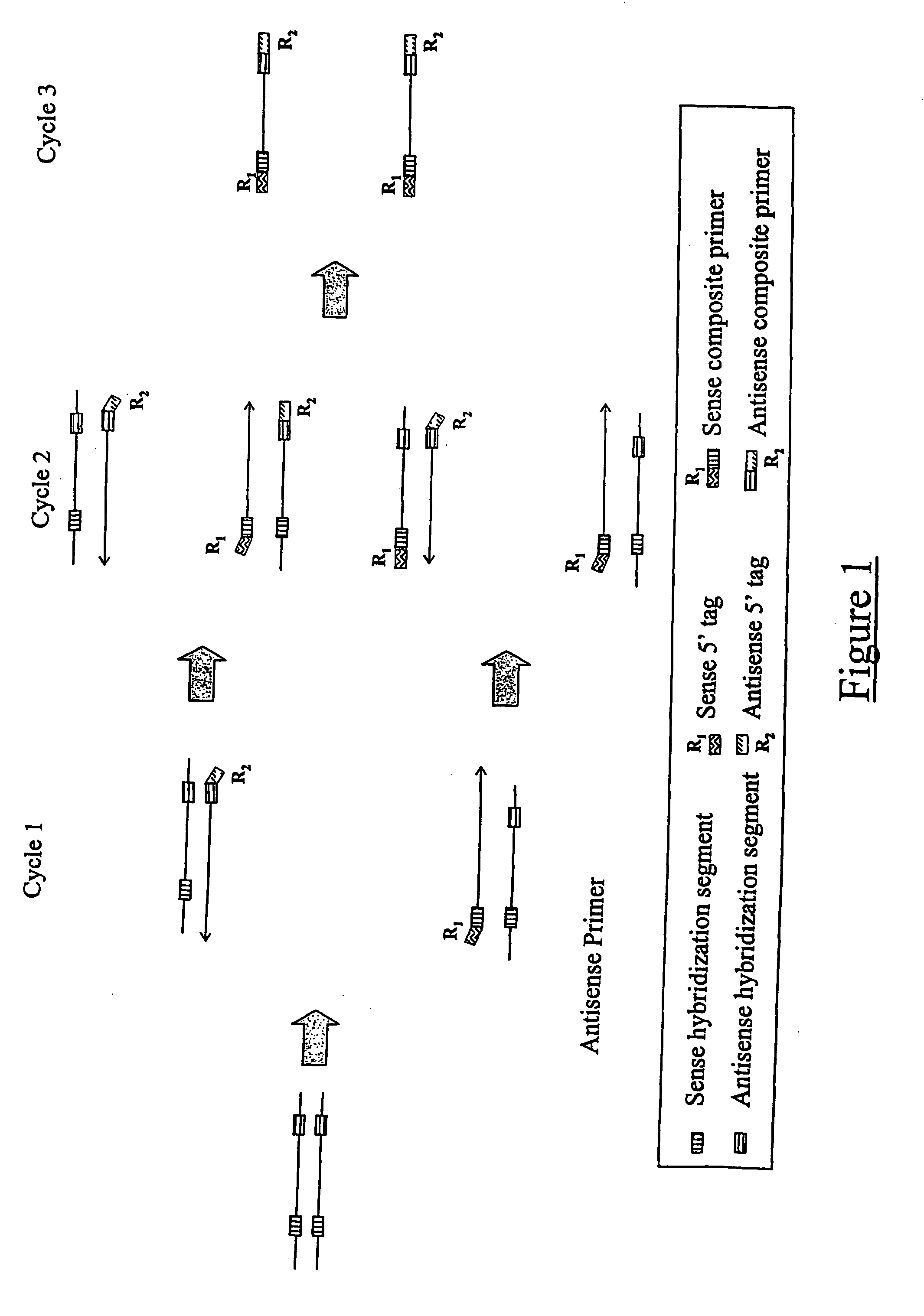
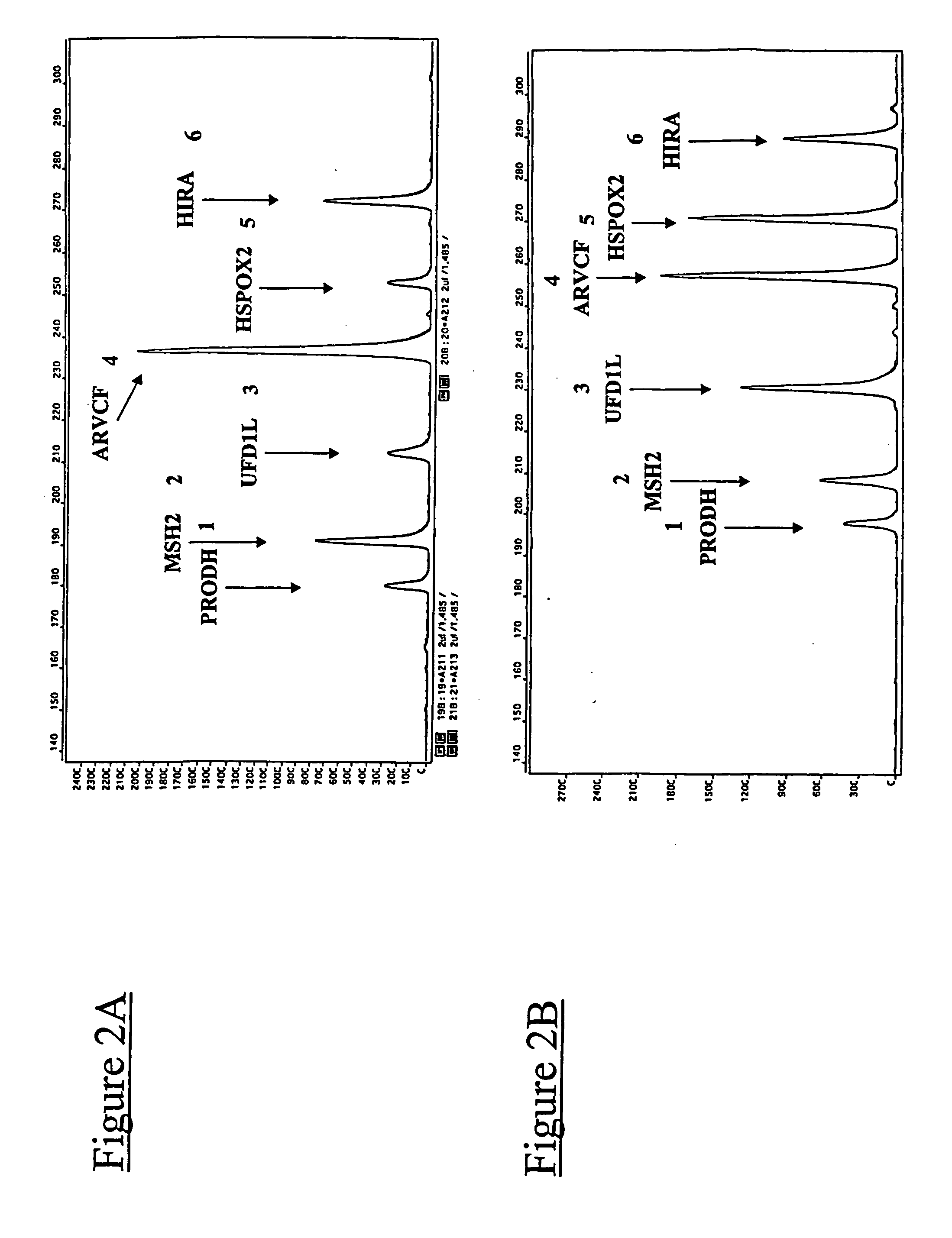
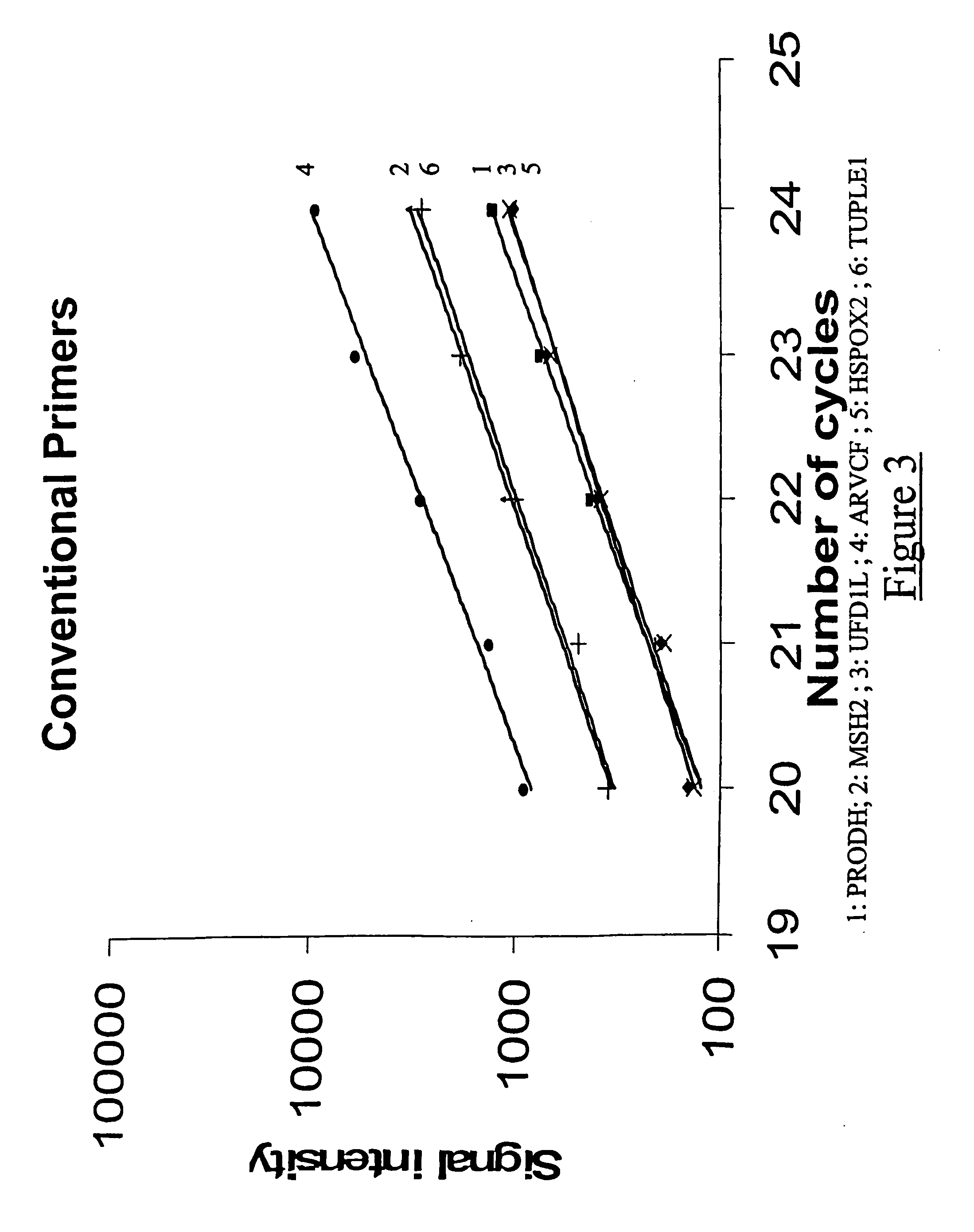
The present application relates to novel composite primers which make it possible to amplify in multiplex at a quantitative level of precision, and to the application of these composite primers for detecting gnomic rearrangements in general and cryptic chromosomal rearrangements in particular. These composite primers contain a tag the sequence of which is absent from or poorly represented in the genome analyzed, and which exhibits a very low propensity to form stable pairings. The composite primers, which contain them, make it possible to carry out multiplex amplifications with quantitative precision on the scale of a genome such as the human genome.
Owner:INST NAT DE LA SANTE & DE LA RECHERCHE MEDICALE (INSERM) +1
Methods and compositions for enrichment of amplification products
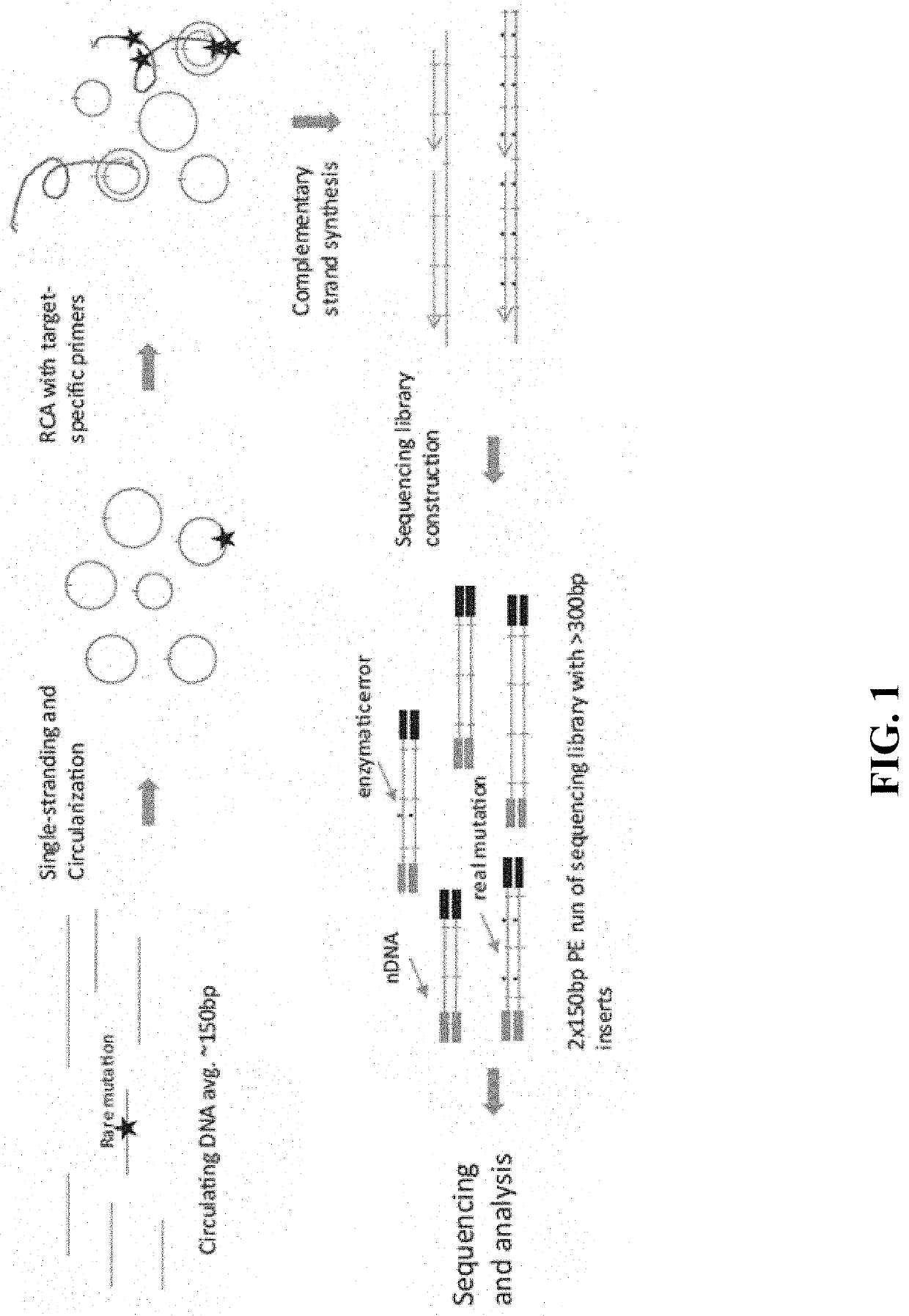
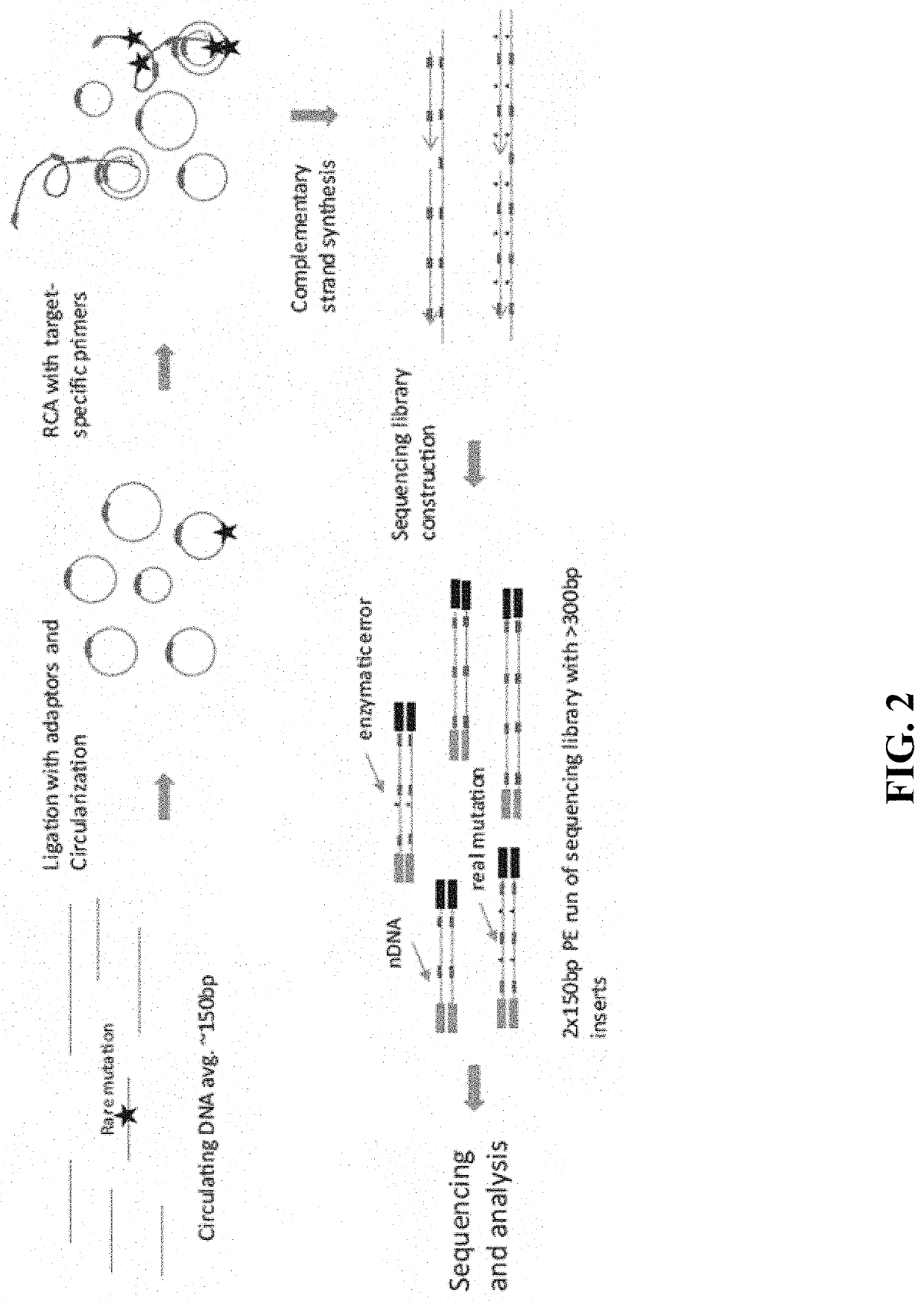
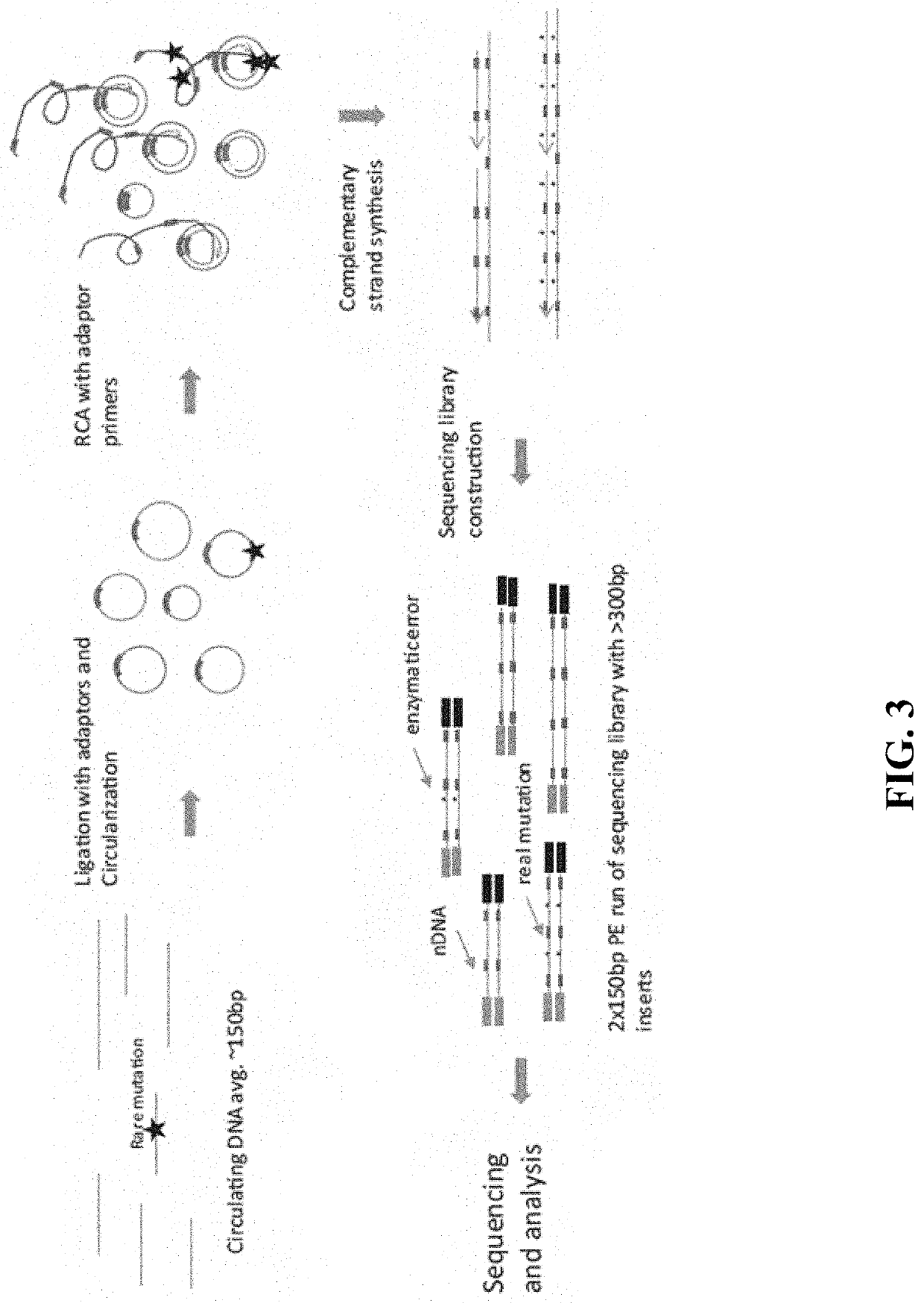
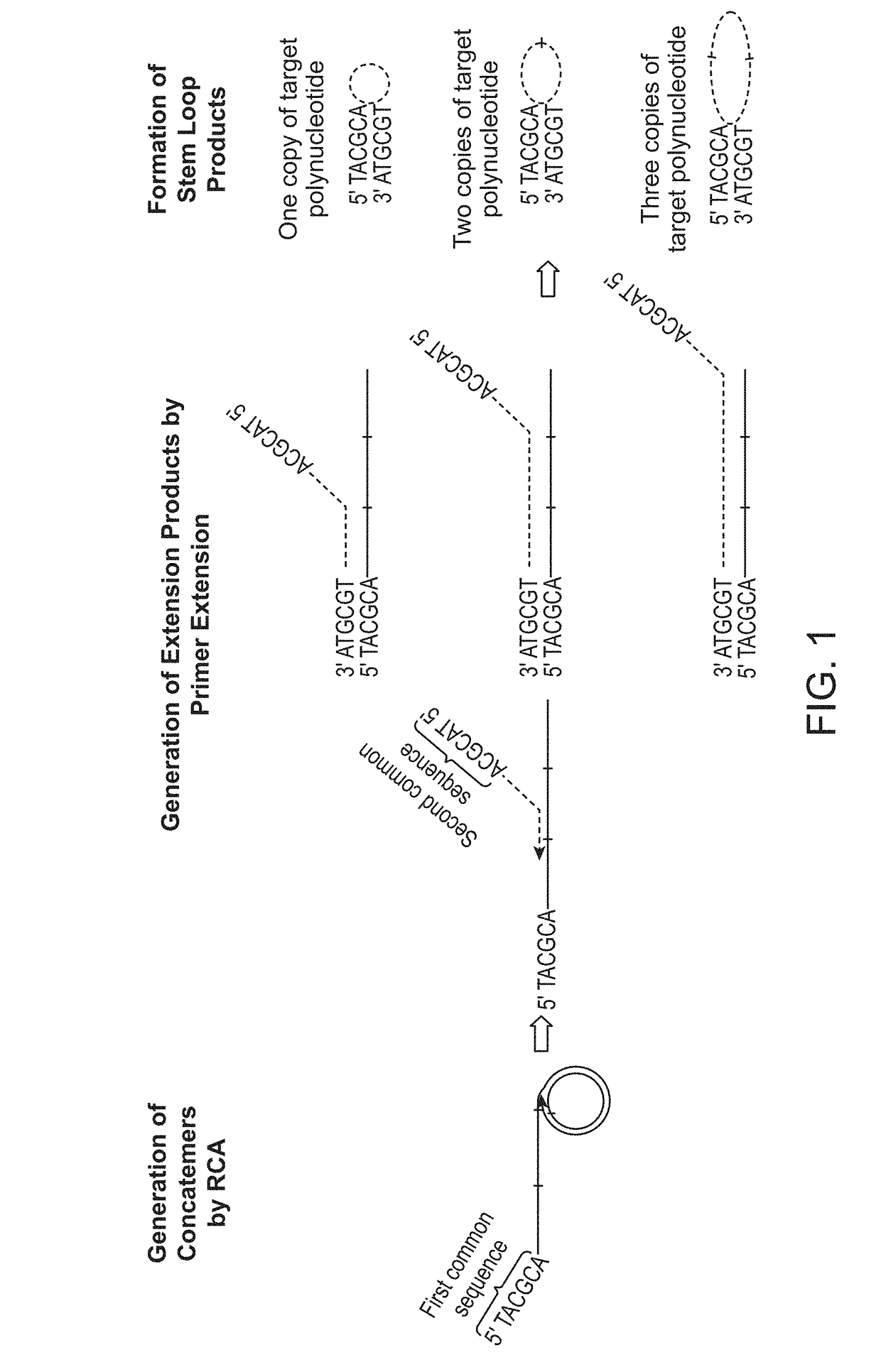
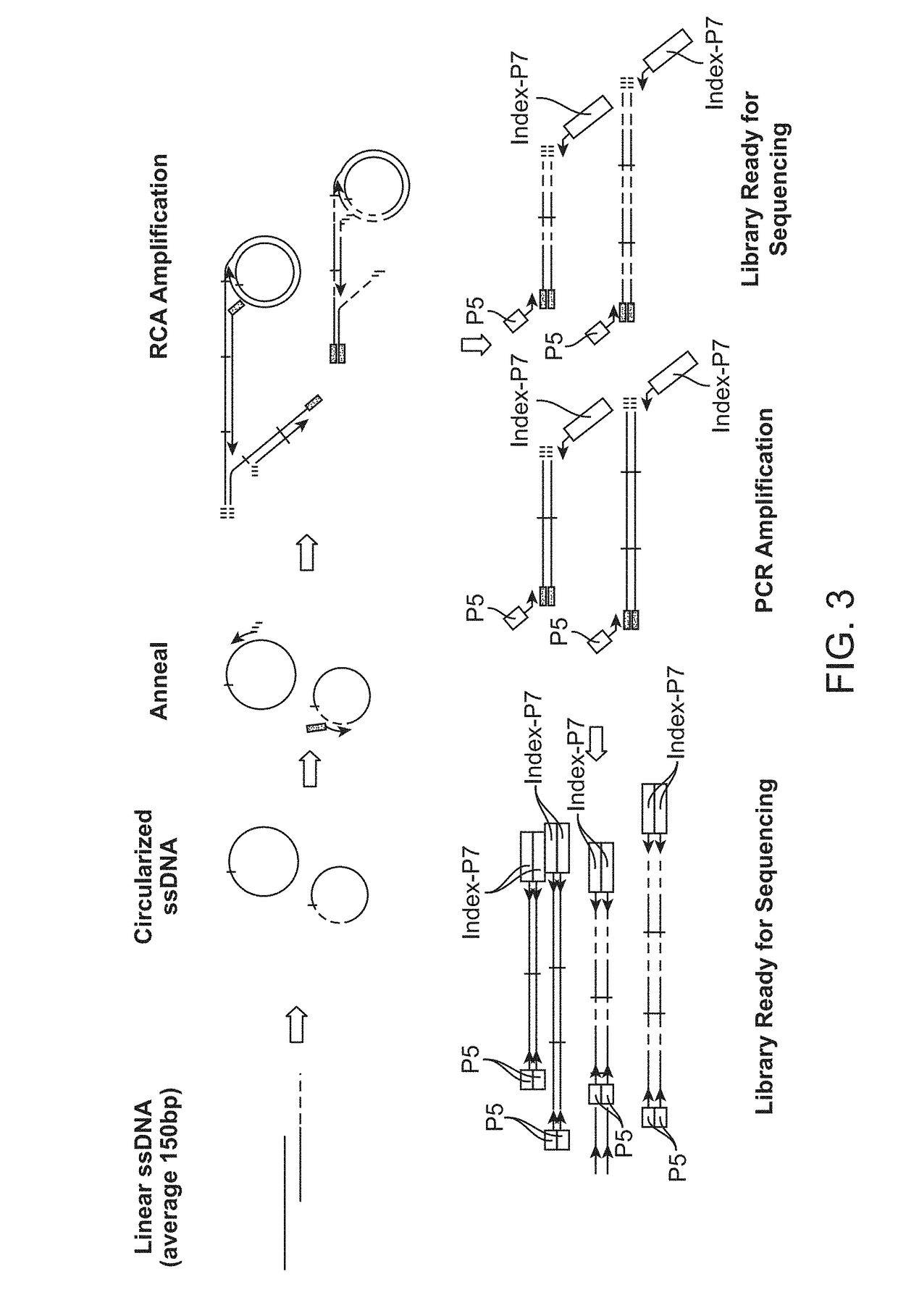
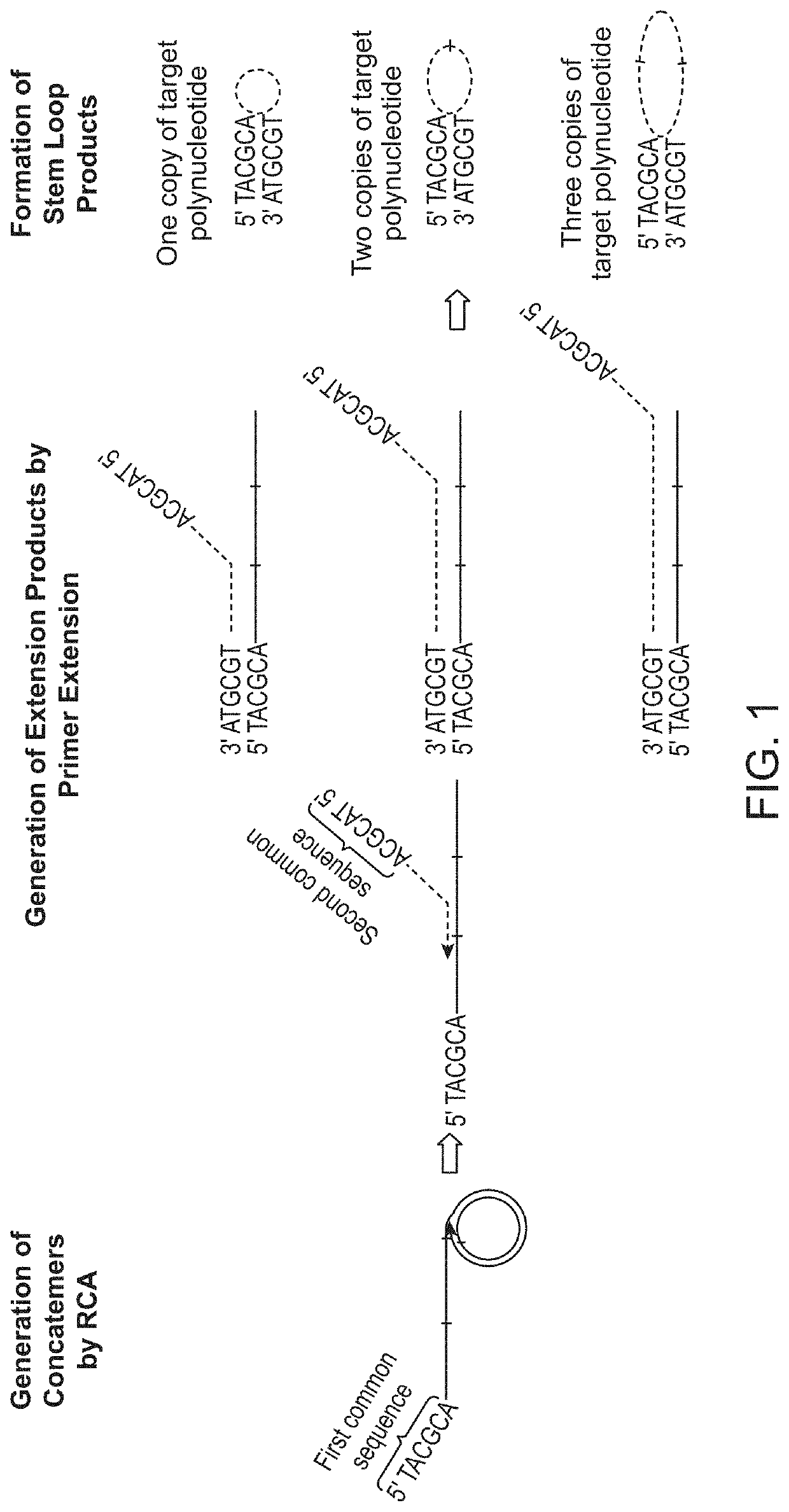
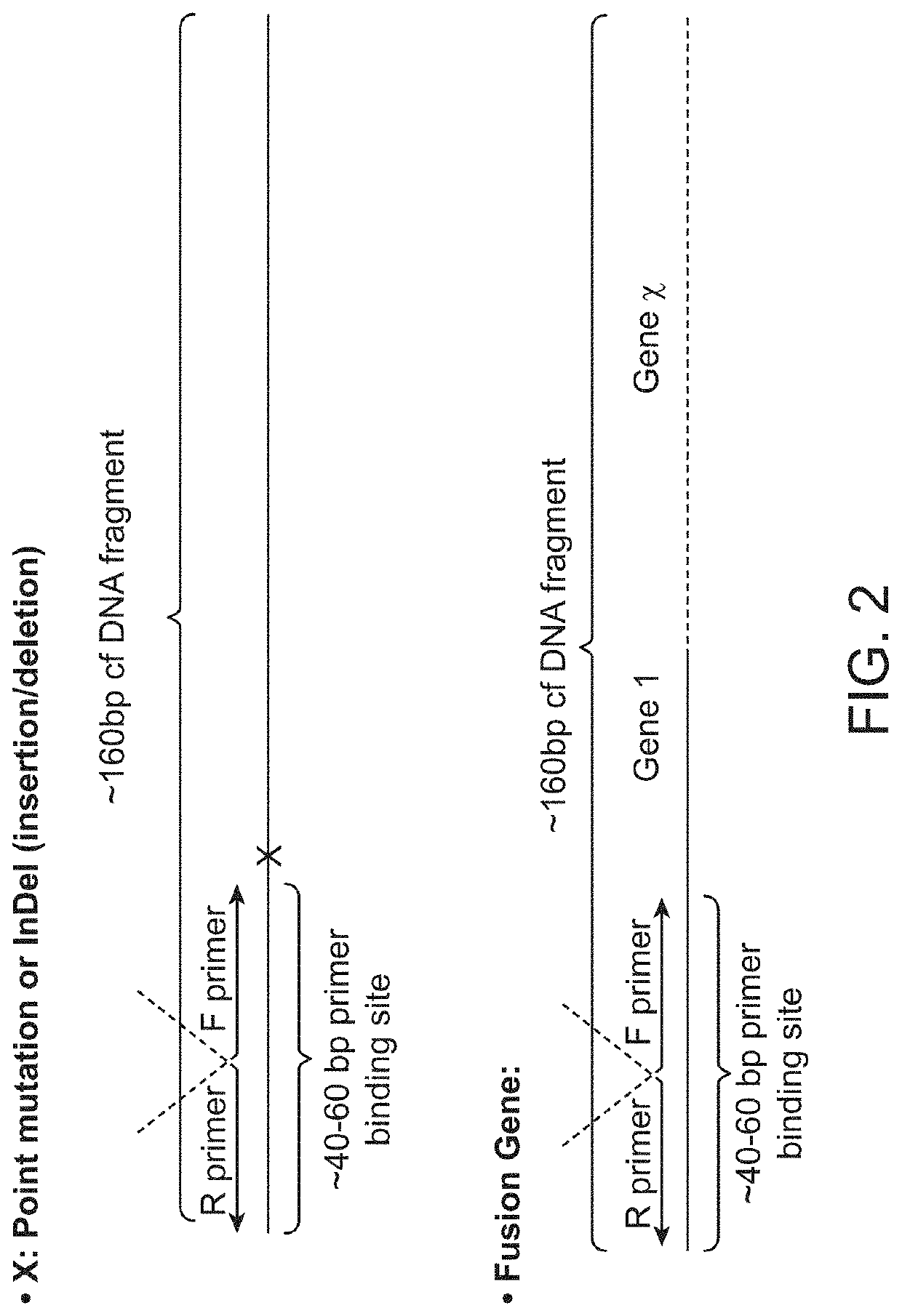
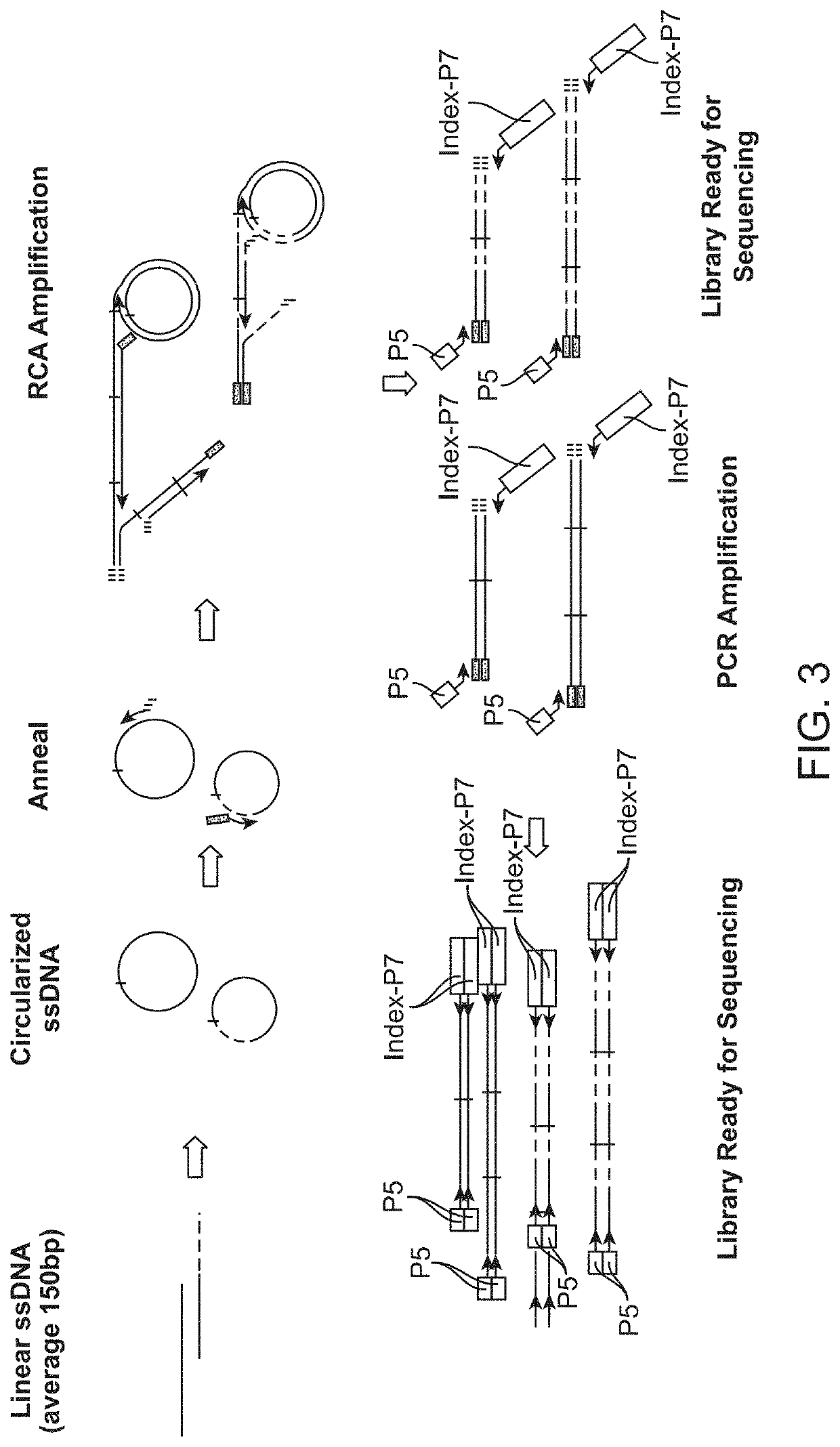
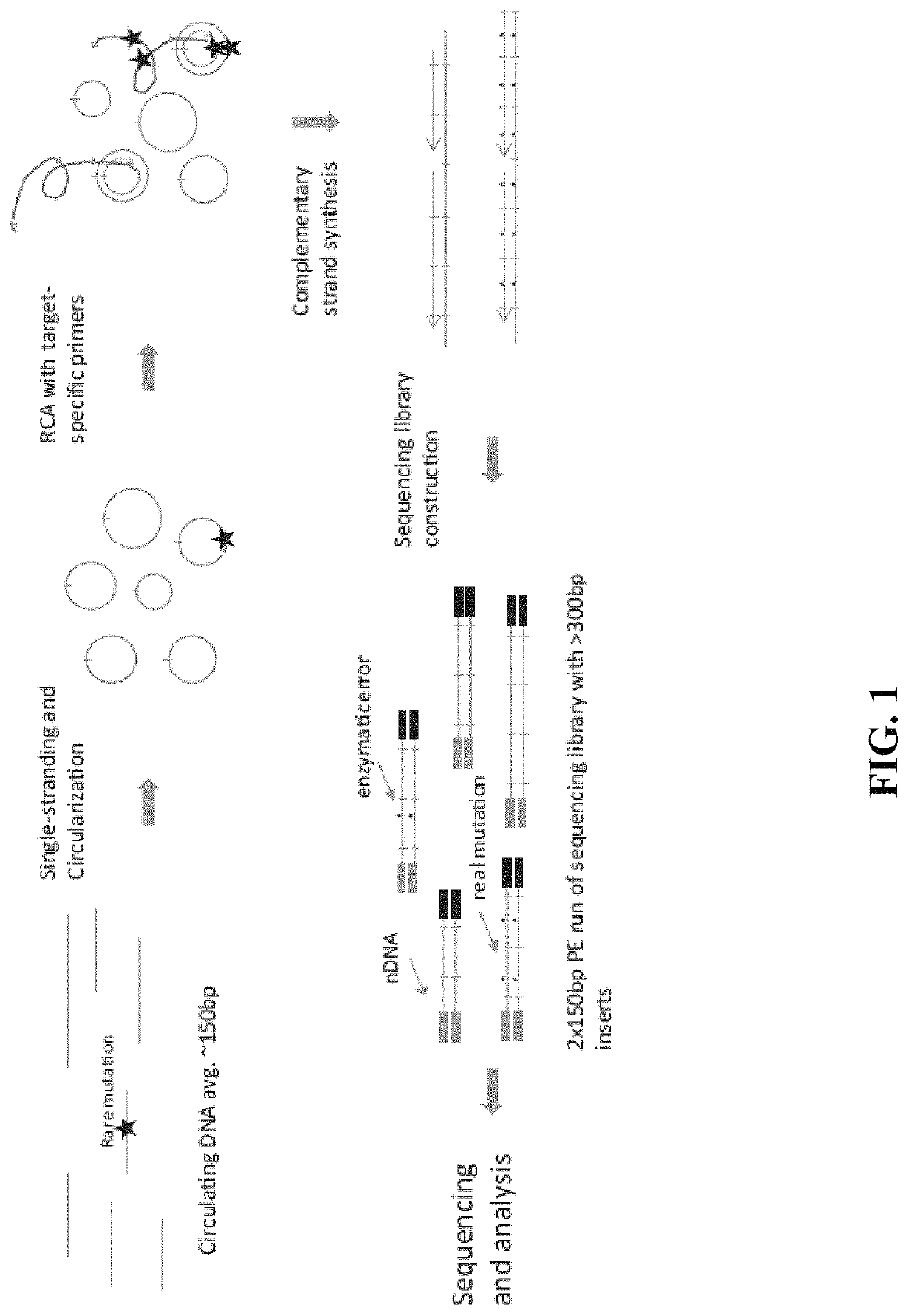
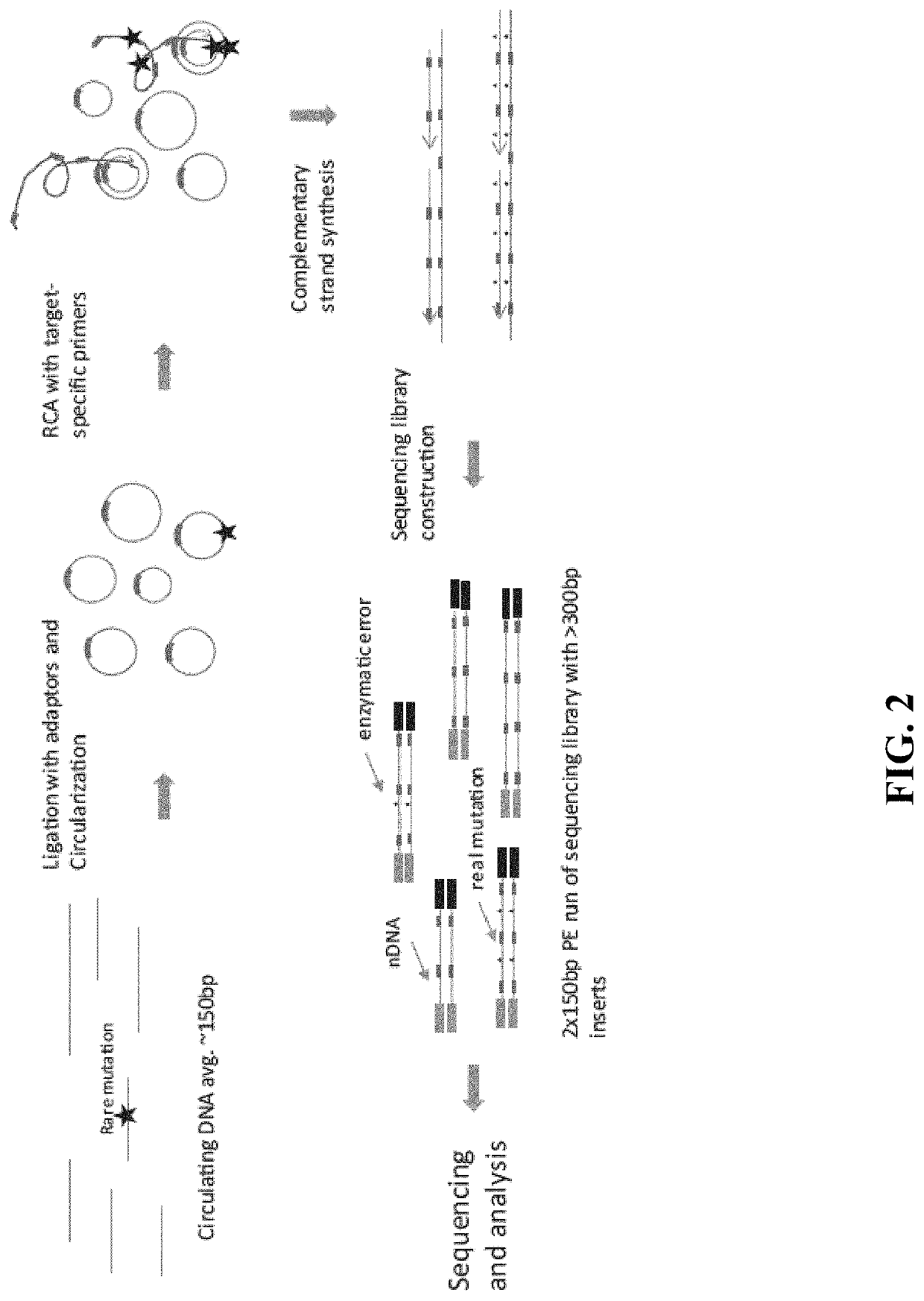
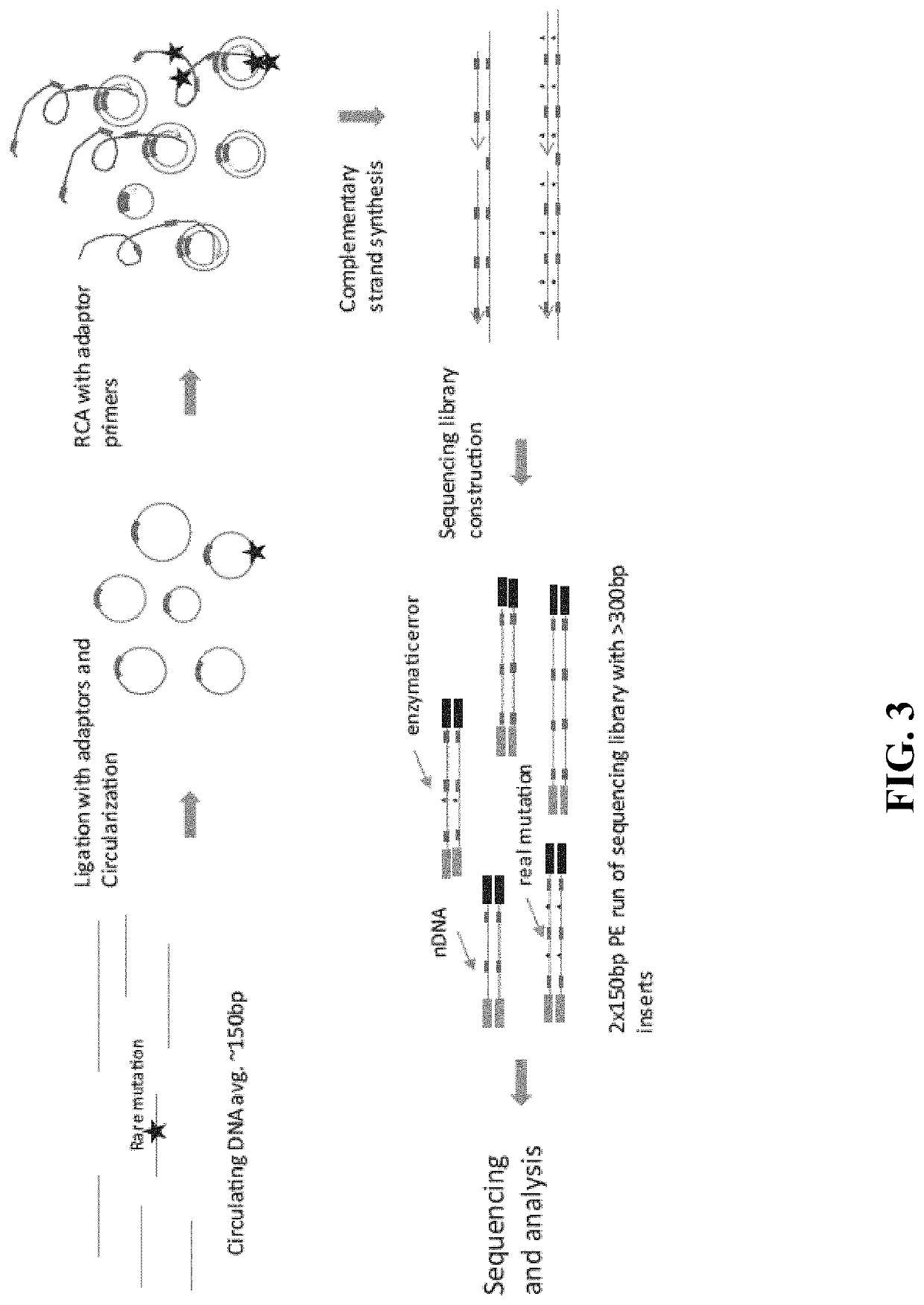
In some aspects, the present disclosure provides methods for enriching amplicons, or amplification products, comprising a concatemer of at least two or more copies of a target polynucleotide. In some embodiments, a method comprises sequencing the amplicons comprising at least two or more copies of a target polynucleotide. In some embodiments, the target polynucleotides comprise sequences resulting from chromosome rearrangement, including but not limited to point mutations, single nucleotide polymorphisms, insertions, deletions, and translocations including fusion genes. In some aspects, the present disclosure provides compositions and reaction mixtures useful in the described methods.
Owner:ACCURAGEN HLDG LTD
Methods and compositions for enrichment of amplification products
ActiveUS20180298434A1Reduce errorsRaise the ratioMicrobiological testing/measurementHybridisationNucleotidePolynucleotide
In some aspects, the present disclosure provides methods for enriching amplicons, or amplification products, comprising a concatemer of at least two or more copies of a target polynucleotide. In some embodiments, a method comprises sequencing the amplicons comprising at least two or more copies of a target polynucleotide. In some embodiments, the target polynucleotides comprise sequences resulting from chromosome rearrangement, including but not limited to point mutations, single nucleotide polymorphisms, insertions, deletions, and translocations including fusion genes. In some aspects, the present disclosure provides compositions and reaction mixtures useful in the described methods.
Owner:ACCURAGEN HLDG LTD
Methods and compositions for enrichment of amplification products
ActiveUS10752942B2Reduce errorsRaise the ratioSugar derivativesMicrobiological testing/measurementNucleotidePolynucleotide
In some aspects, the present disclosure provides methods for enriching amplicons, or amplification products, comprising a concatemer of at least two or more copies of a target polynucleotide. In some embodiments, a method comprises sequencing the amplicons comprising at least two or more copies of a target polynucleotide. In some embodiments, the target polynucleotides comprise sequences resulting from chromosome rearrangement, including but not limited to point mutations, single nucleotide polymorphisms, insertions, deletions, and translocations including fusion genes. In some aspects, the present disclosure provides compositions and reaction mixtures useful in the described methods.
Owner:ACCURAGEN HLDG LTD
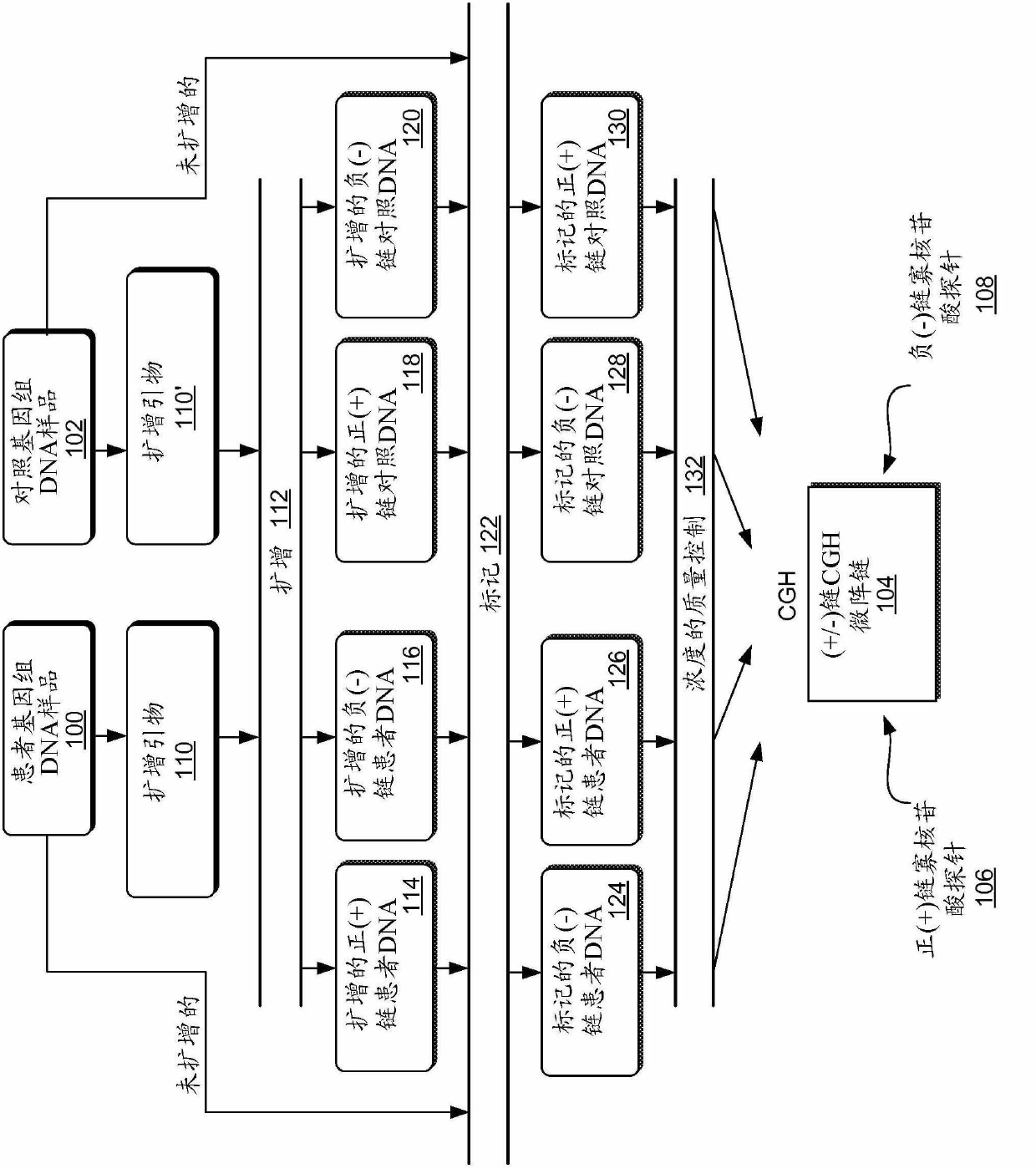
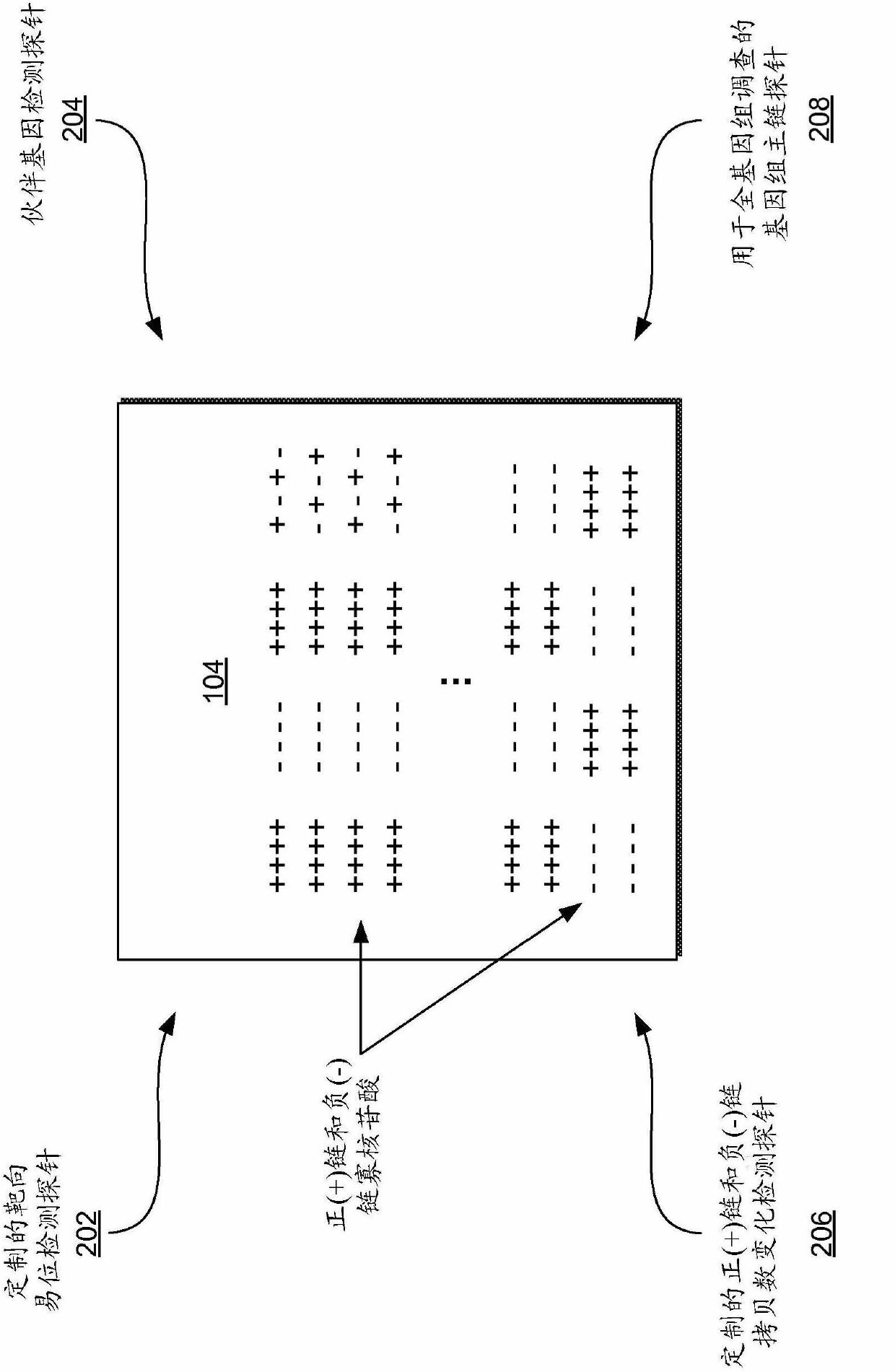
Multiplex (+/-) stranded arrays and assays for detecting chromosomal abnormalities associated with cancer and other diseases
InactiveCN102630250AMicrobiological testing/measurementArray-Based Comparative Genomic HybridizationProtein translocation
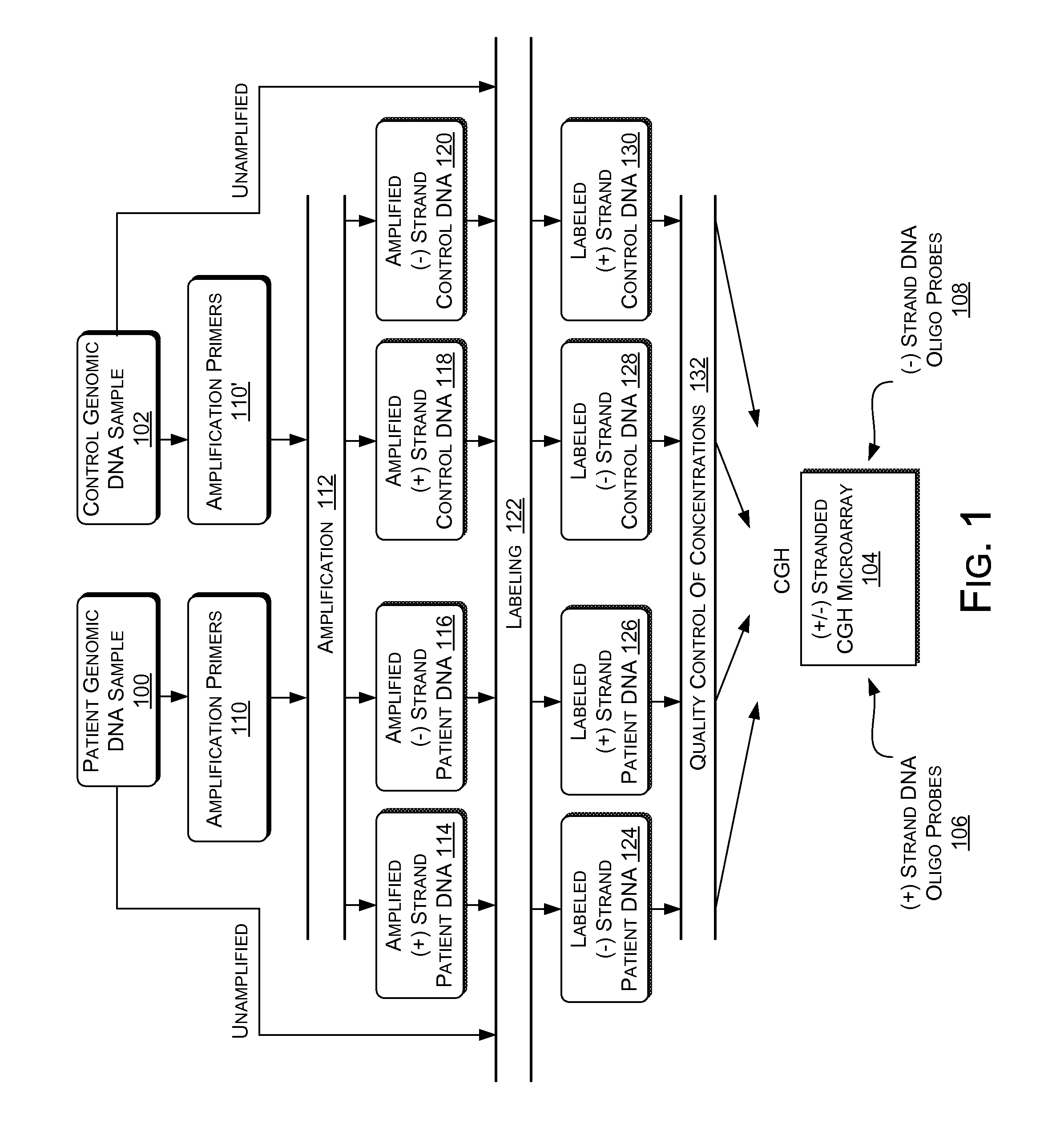
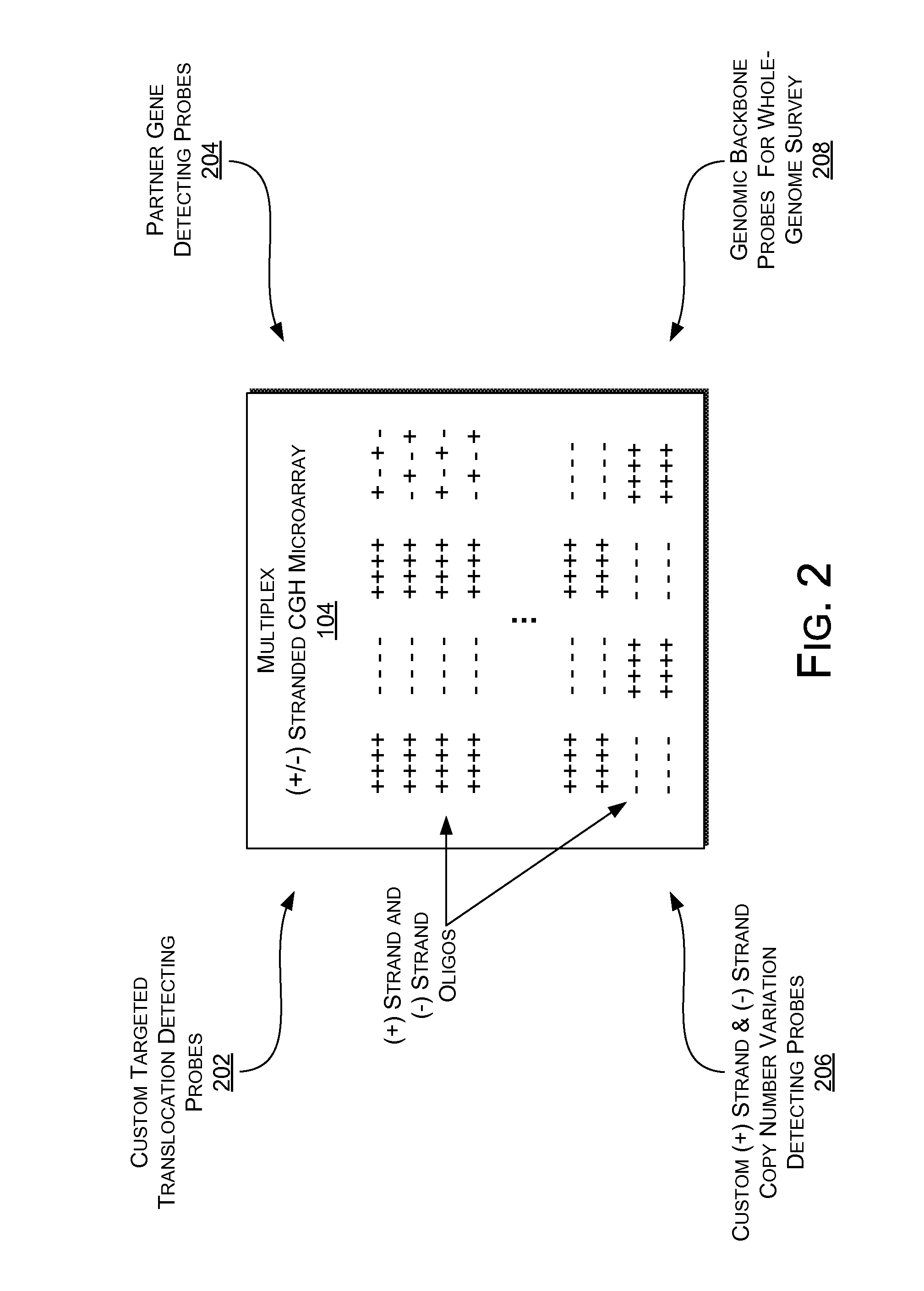
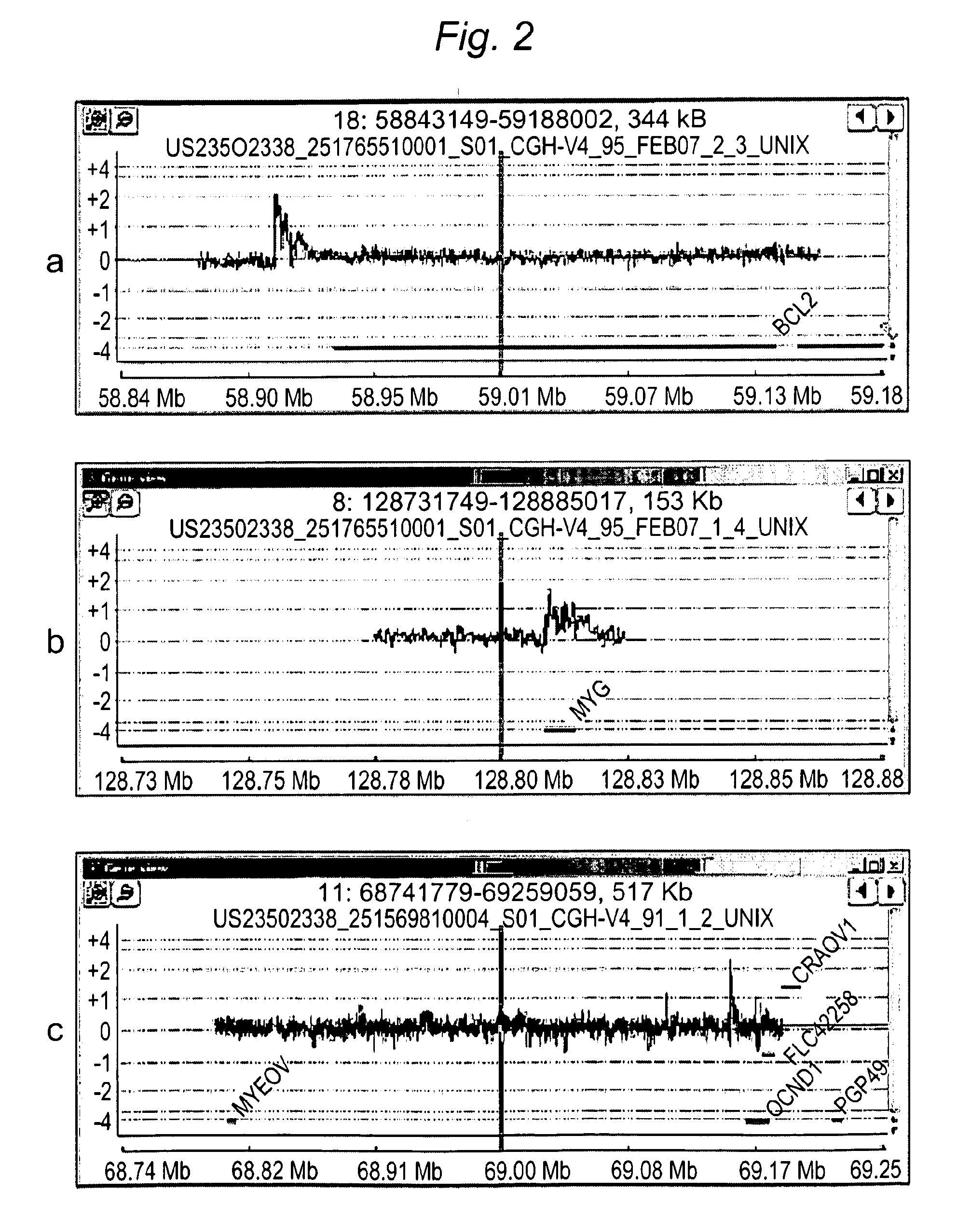
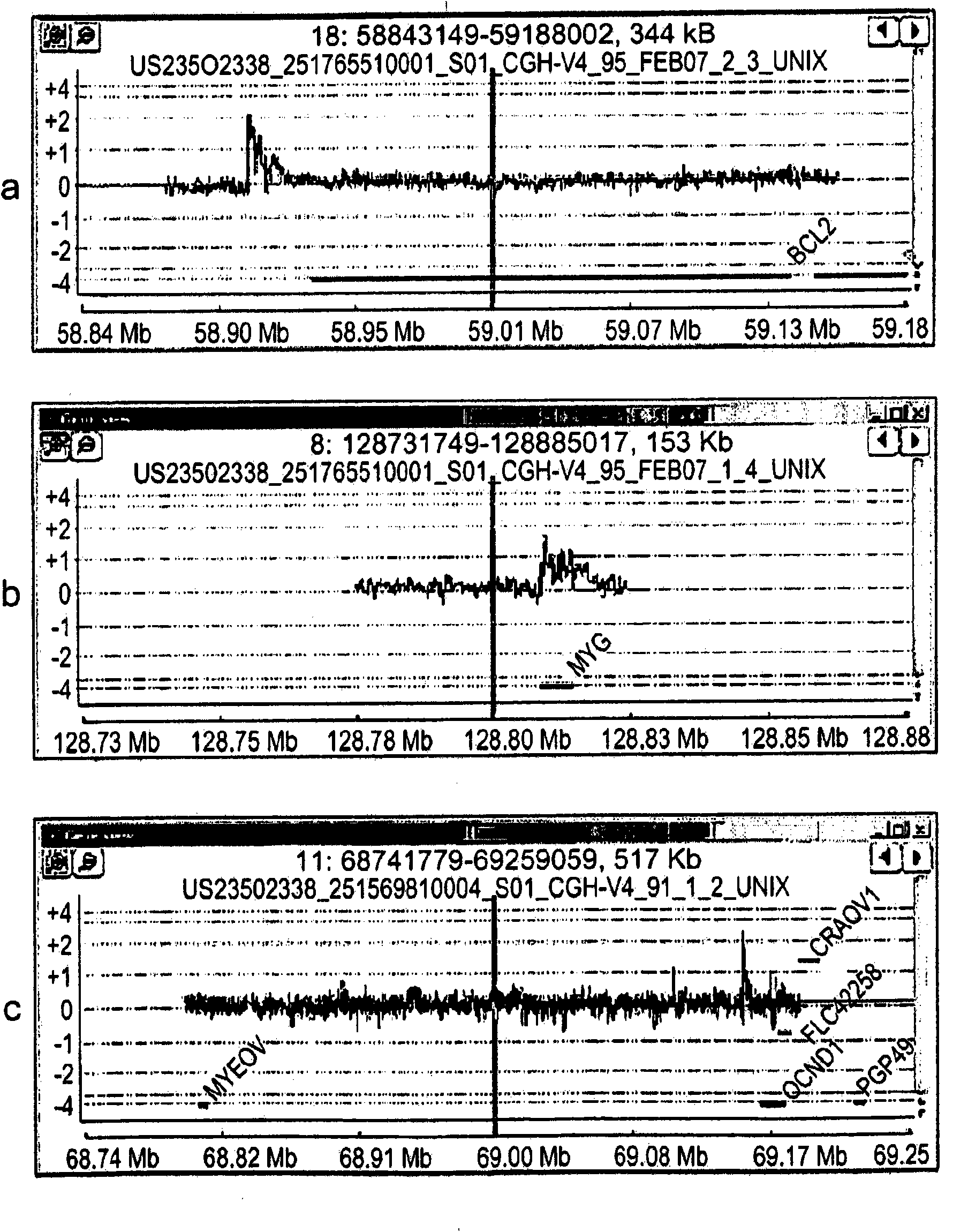
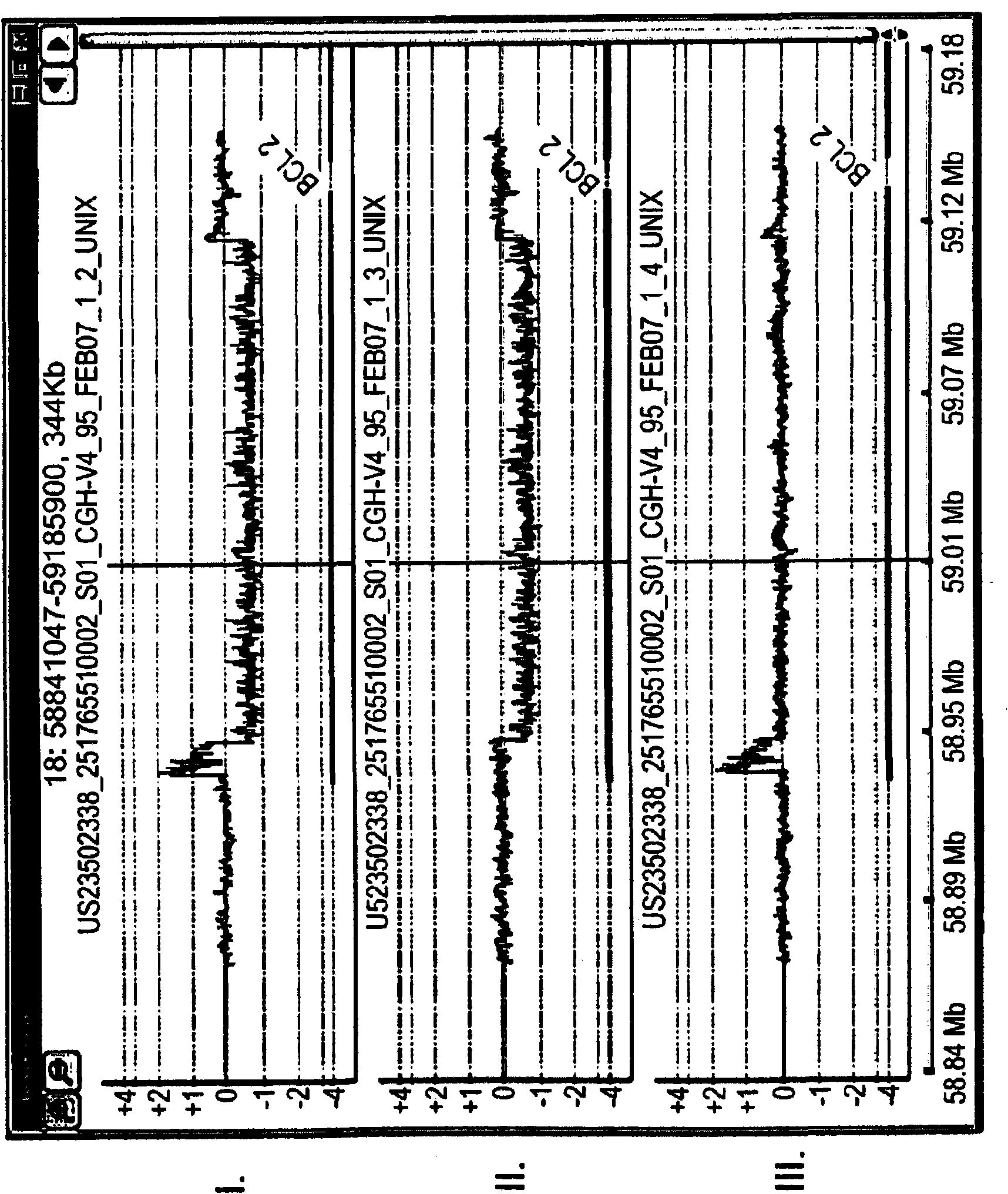
Multiplex (+ / -) stranded analyses, such as array comparative genomic hybridization (aCGH), are provided for detecting chromosomal rearrangements associated with cancer and other diseases. For example, an illustrative multiplex array for CGH includes discrete plus (+) strand and minus (-) strand DNA probes, complementary to each other but separable on the CGH array. The minus (-) strand DNA probes recover diagnostic information lost to conventional microarrays, since many genes transcribe from the minus (-) strand. In an illustrative system, patient and control DNA samples are prepared for CGH by amplification and labeling using comprehensive primers that generate both plus (+) strands and minus (-) strands of DNA in the samples. The breakpoints of a translocated chromosome may be detected on a multiplex microarray by DNA probes of one polarity, while DNA copy number changes associated with the translocation region may be detected by corresponding DNA probes of the complementary polarity. Related methods for identifying translocation partner genes are also provided.
Owner:SIGNATURE GENOMICS LAB
Algorithm for modification of somatic cancer evolution
Owner:RUBBEN ALBERT
Multiplex (+/-) stranded arrays and assays for detecting chromosomal abnormalities associated with cancer and other diseases
InactiveUS20110086772A1Comprehensive and accurate profile signatureMicrobiological testing/measurementLibrary screeningDiagnostic informationArray-Based Comparative Genomic Hybridization
Multiplex (+ / −) stranded analyses, such as array comparative genomic hybridization (aCGH), are provided for detecting chromosomal rearrangements associated with cancer and other diseases. For example, an illustrative multiplex array for CGH includes discrete plus (+) strand and minus (−) strand DNA probes, complementary to each other but separable on the CGH array. The minus (−) strand DNA probes recover diagnostic information lost to conventional microarrays, since many genes transcribe from the minus (−) strand. In an illustrative system, patient and control DNA samples are prepared for CGH by amplification and labeling using comprehensive primers that generate both plus (+) strands and minus (−) strands of DNA in the samples. The breakpoints of a translocated chromosome may be detected on a multiplex microarray by DNA probes of one polarity, while DNA copy number changes associated with the translocation region may be detected by corresponding DNA probes of the complementary polarity. Related methods for identifying translocation partner genes are also provided.
Owner:SIGNATURE GENOMICS LAB
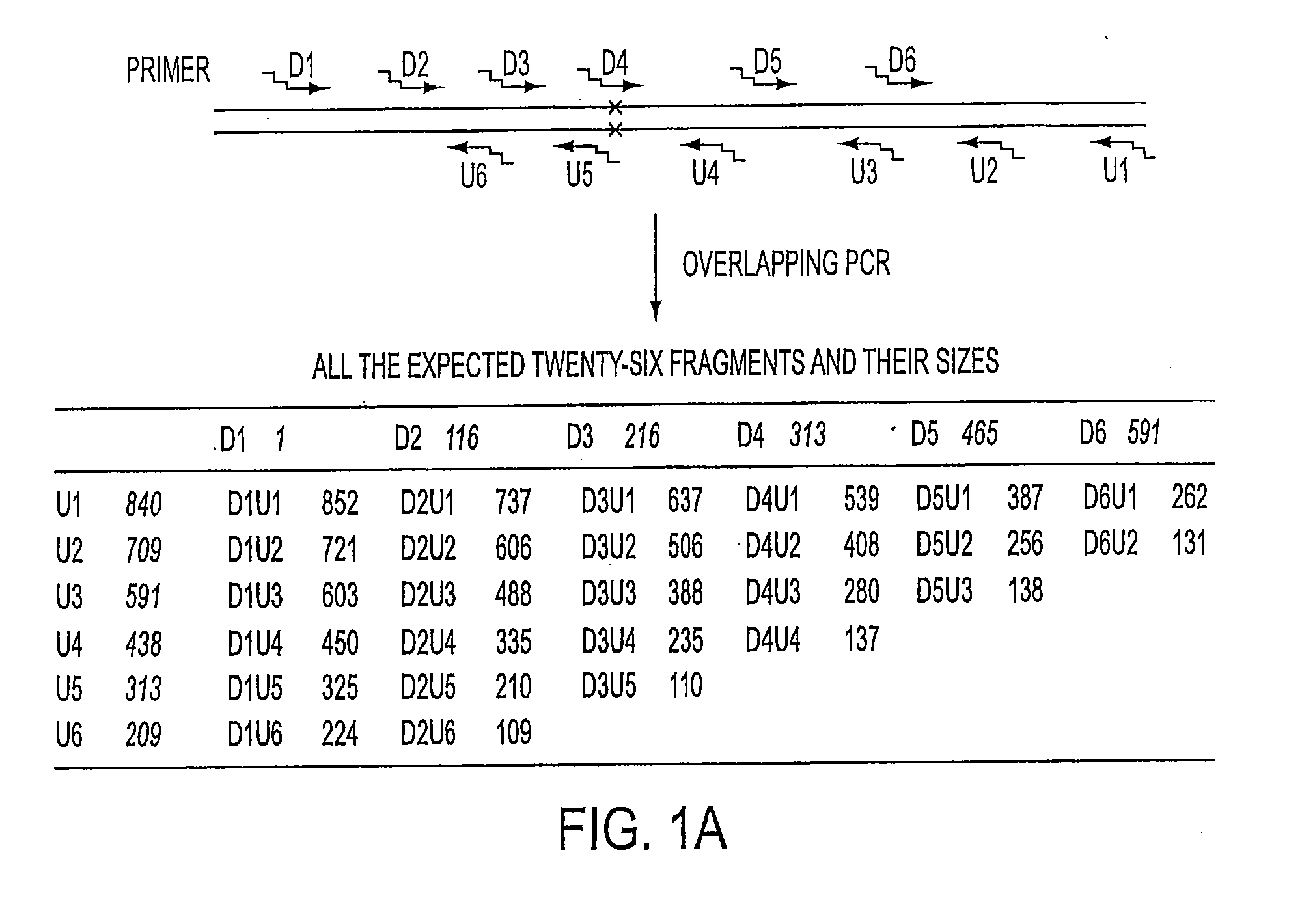
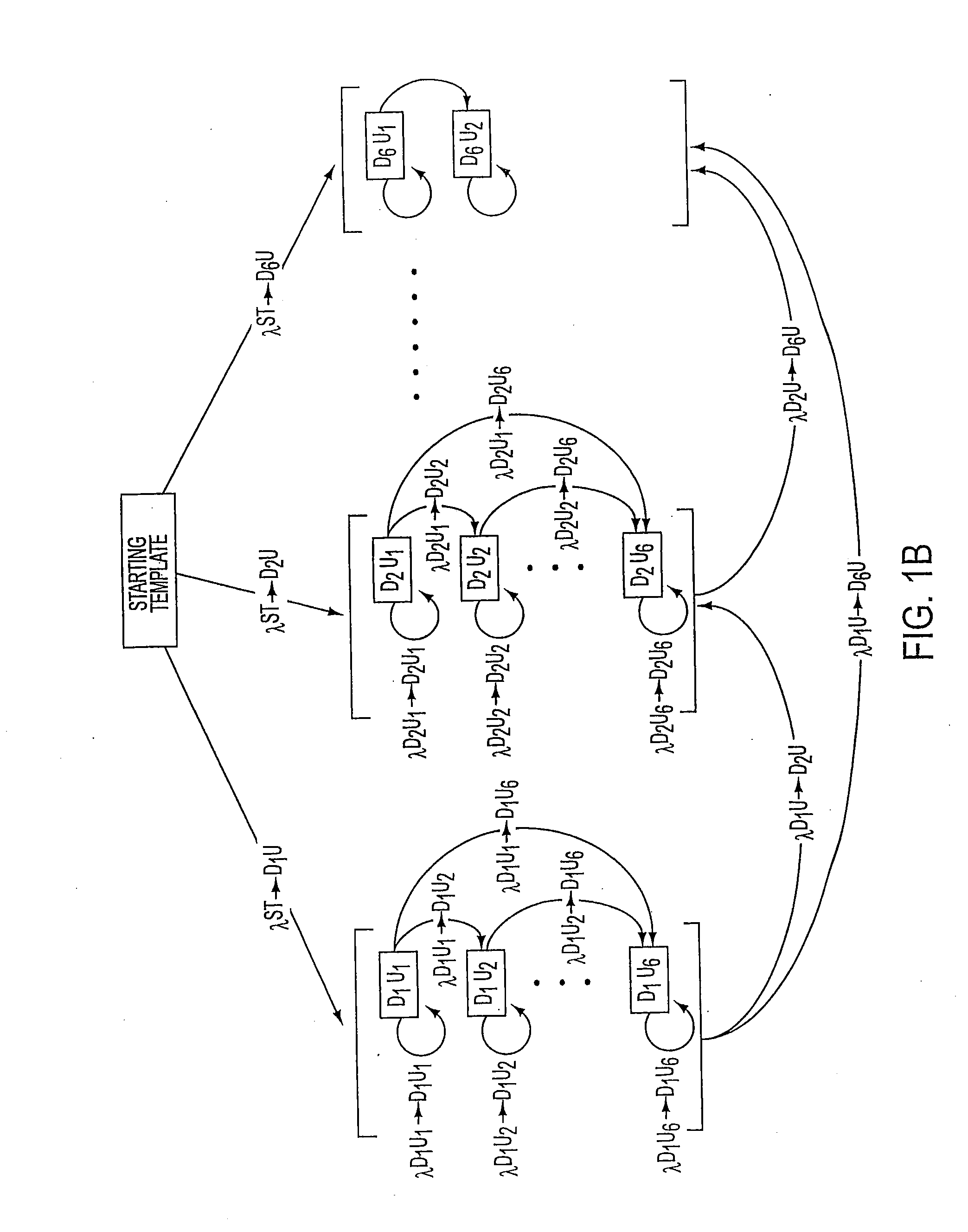
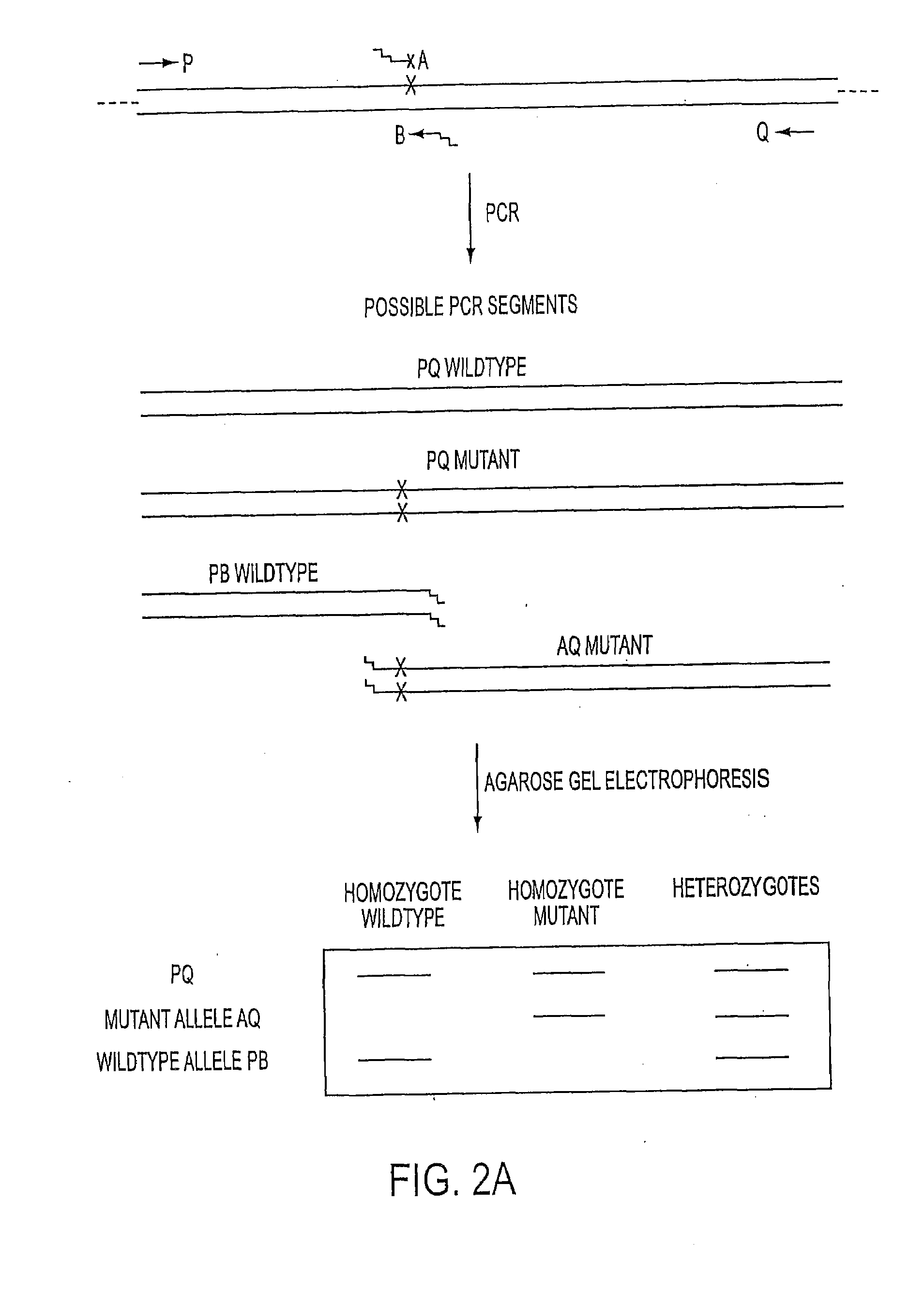
Long distance polymerase chain reaction-based assay for detecting chromosomal rearrangements
Methods are presented for determining the presence of an inversion in the factor VIII gene which cause hemophilia A. The methods encompass long distance, multiplex PCR (including overlapping PCR). The use of deaza-dGTP, high levels of DNA polymerases and high levels of DMSO aid in successfully performing the PCR. The use of a novel technique called subcycling PCR can also be applied as part of the methods. The technique allows for the determination of whether a person is homozygous or hemizygous for the inversion and has hemophilia A or whether a person is heterozygous for the inversion and is a carrier. The technique of long distance, multiplex PCR including use of deaza-dGTP, high levels of DNA polymerases and high levels of DMSO are applicable to the determination of the presence of other gross chromosomal aberrations such as deletions / inversions, translocations and inversions. The use of subcycling PCR can achieve efficient and more even amplification than normal two or three temperature PCR and is applicable to long distance, multiplex PCR.
Owner:CITY OF HOPE
DNA microarray based identification and mapping of balanced translocation breakpoints
InactiveUS20110021371A1Increase rangeSuper high resolutionMicrobiological testing/measurementLibrary screeningDNA microarrayComparative genomic hybridization
Methods for detecting and mapping chromosomal rearrangements associated with various diseases using comparative genomic hybridization are disclosed. Included are methods to identify translocation partners of known genomic loci and to determine translocation breakpoints. These methods may be used in the prognosis, diagnosis, and determination of predisposition to diseases that involve chromosomal rearrangements.
Owner:UNIV OF WASHINGTON
Presence of erg gene rearrangements and protein over-expression in low grade PIN (LG-PIN) in prostate biopsies
ActiveCN103392009AMicrobiological testing/measurementBiological material analysisChromosomal rearrangementNucleic acid
Owner:VENTANA MEDICAL SYST INC
Algorithm for Modification of Somatic Cancer Evolution
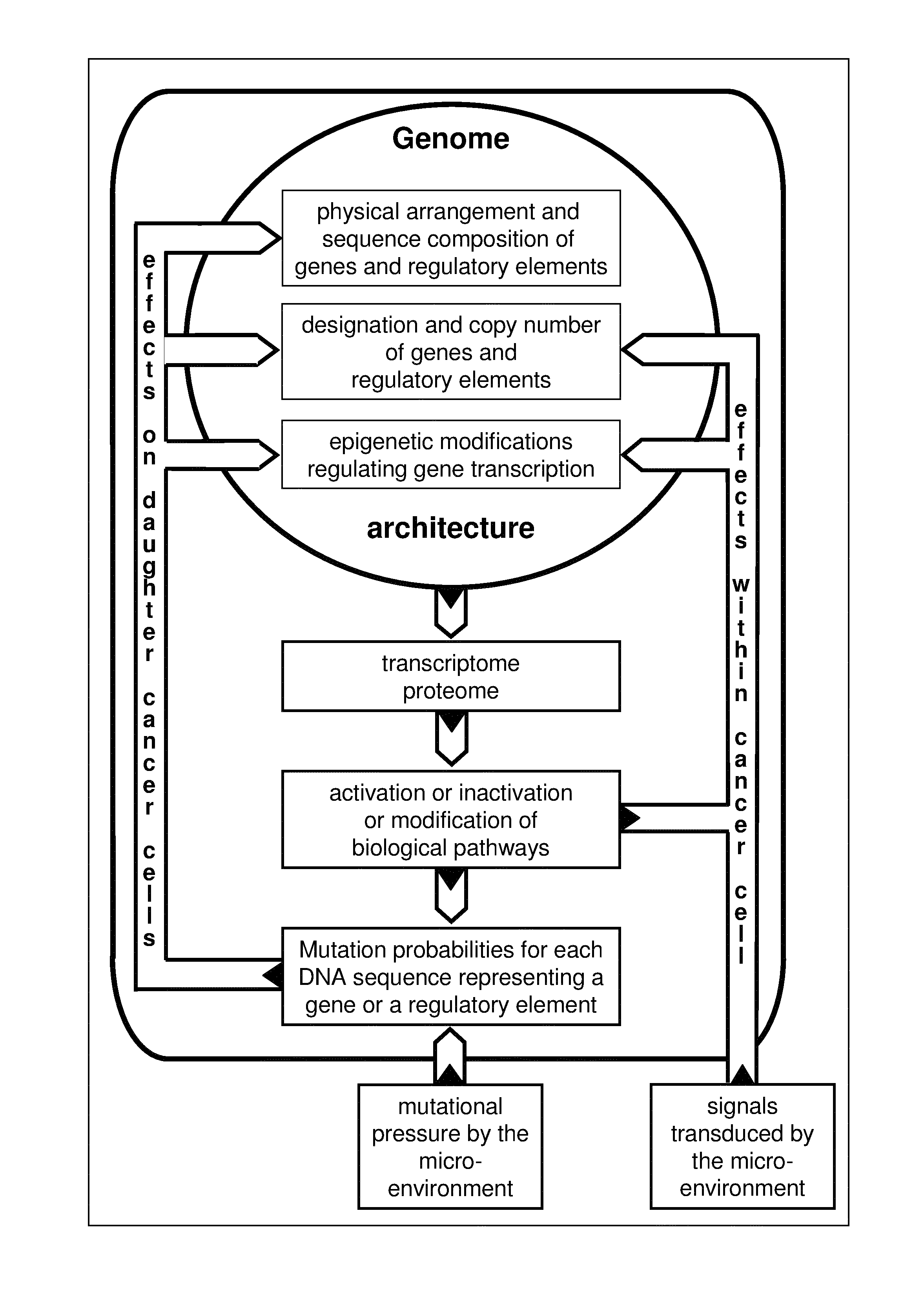
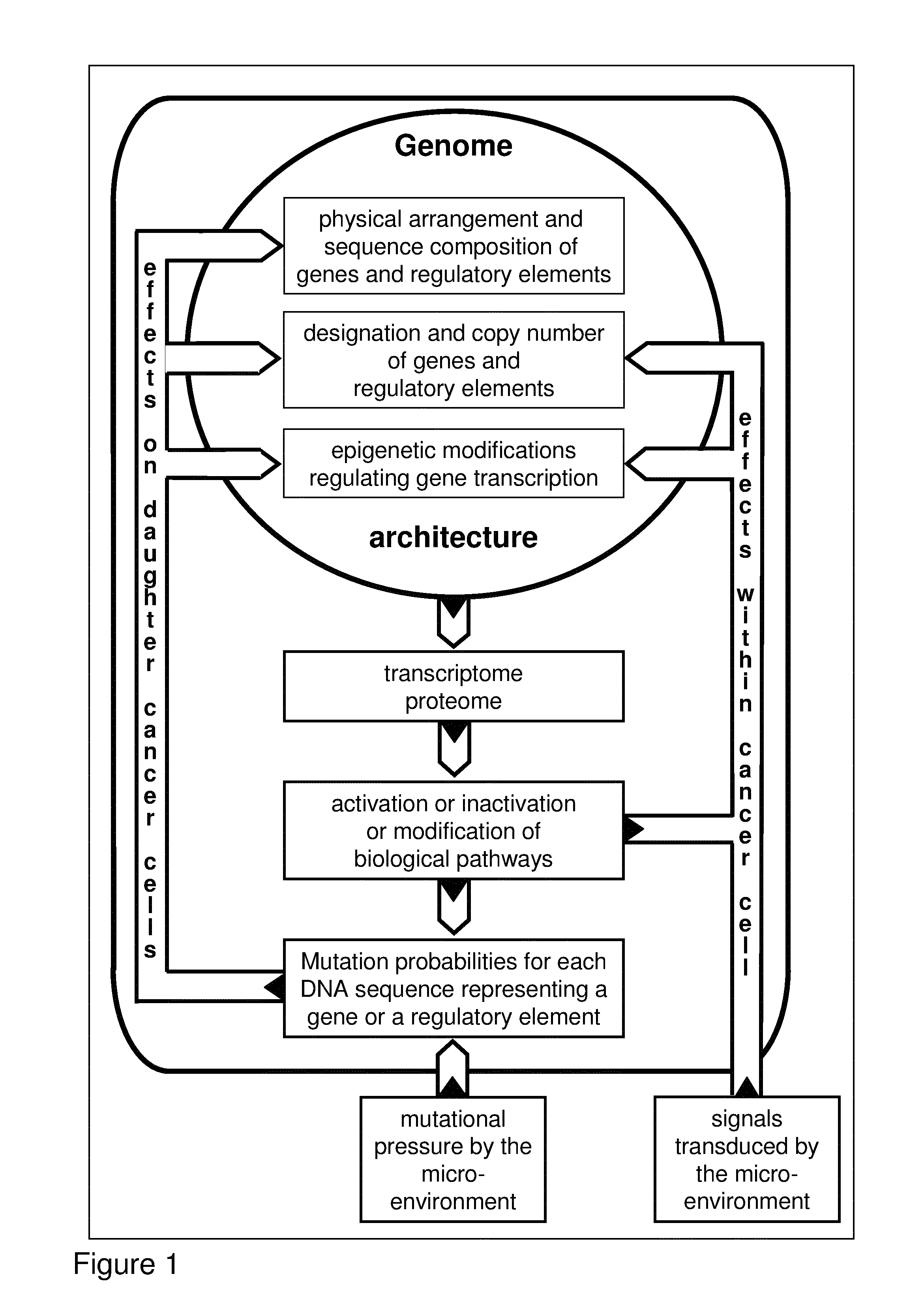
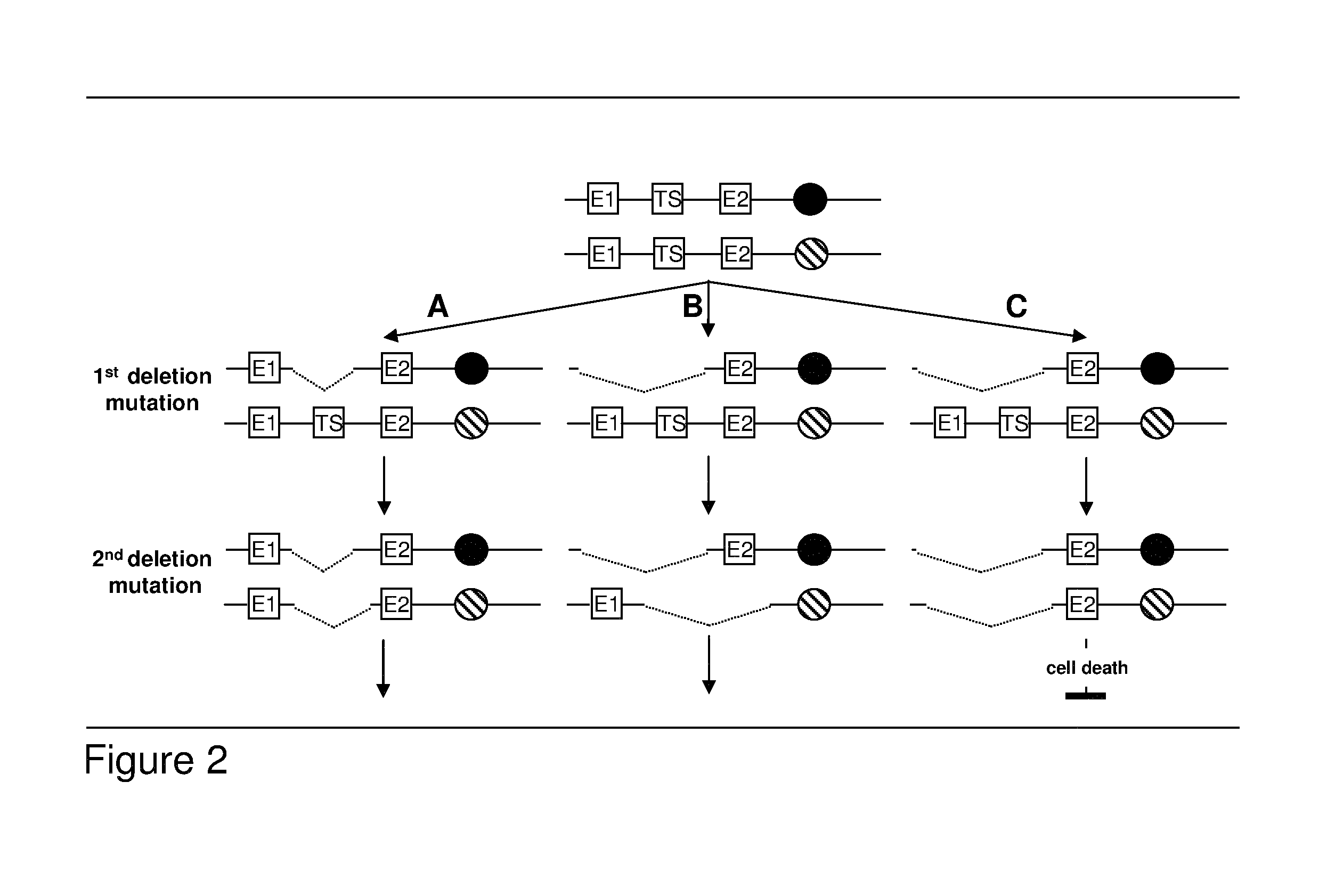
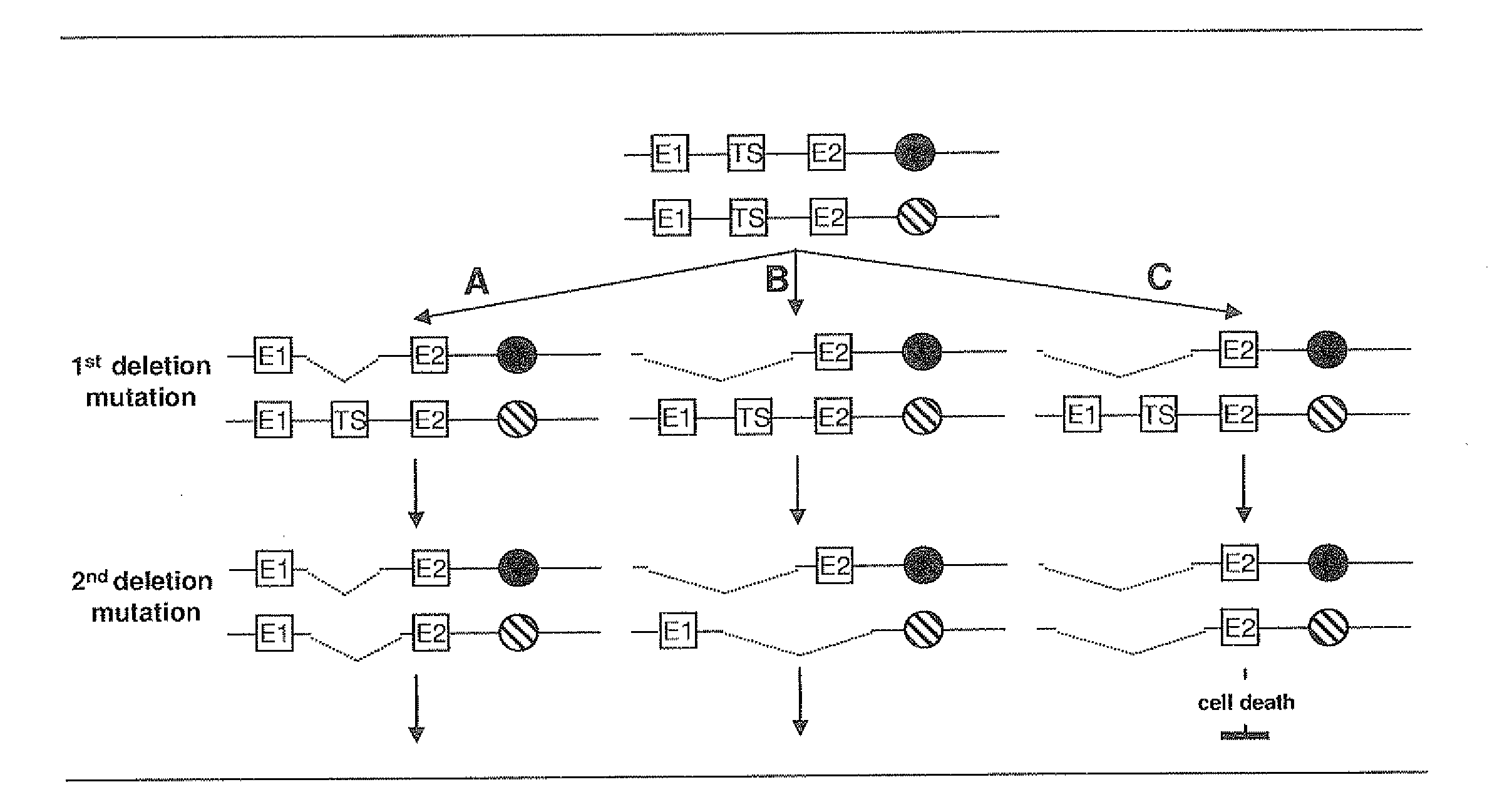
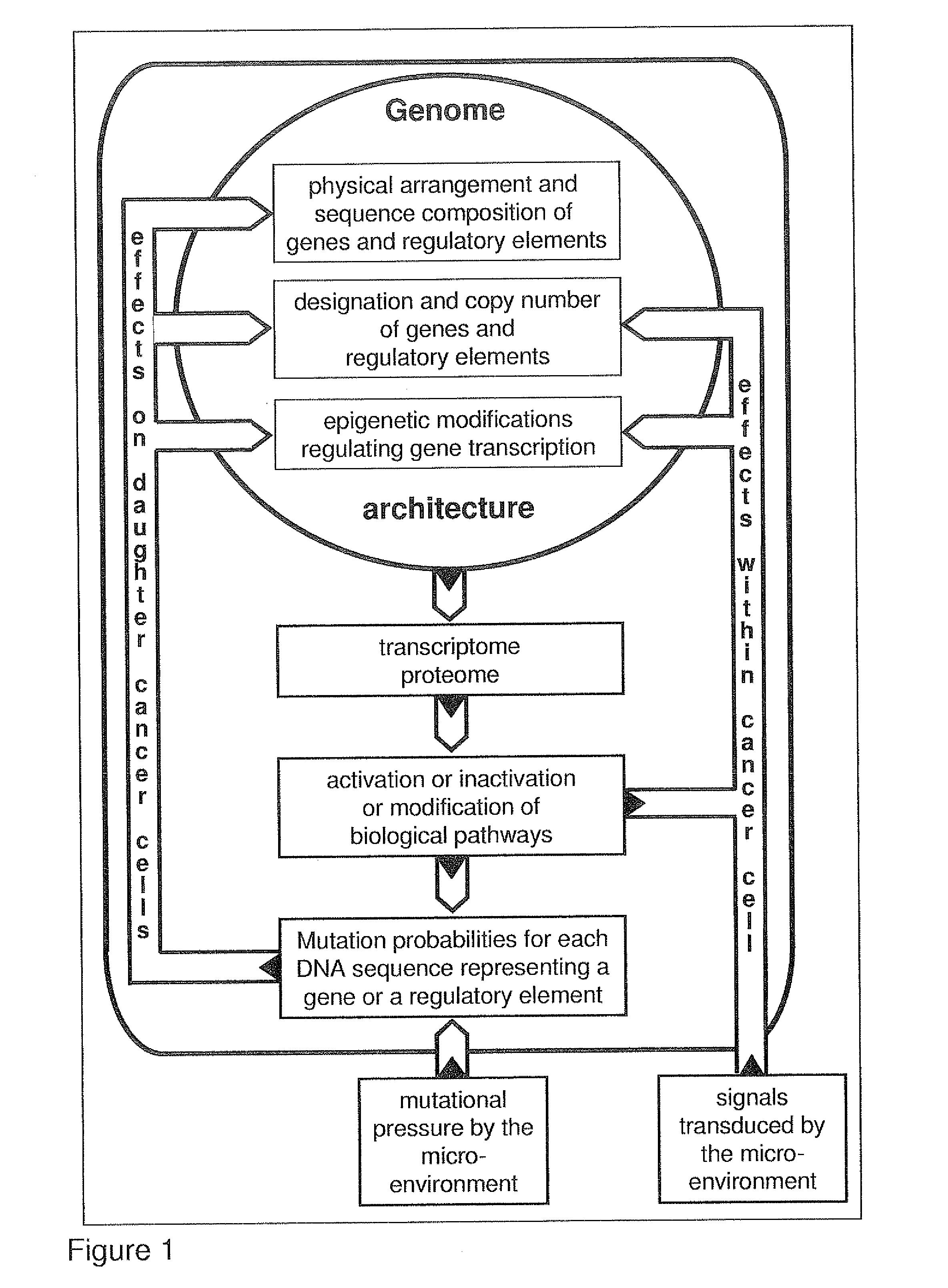
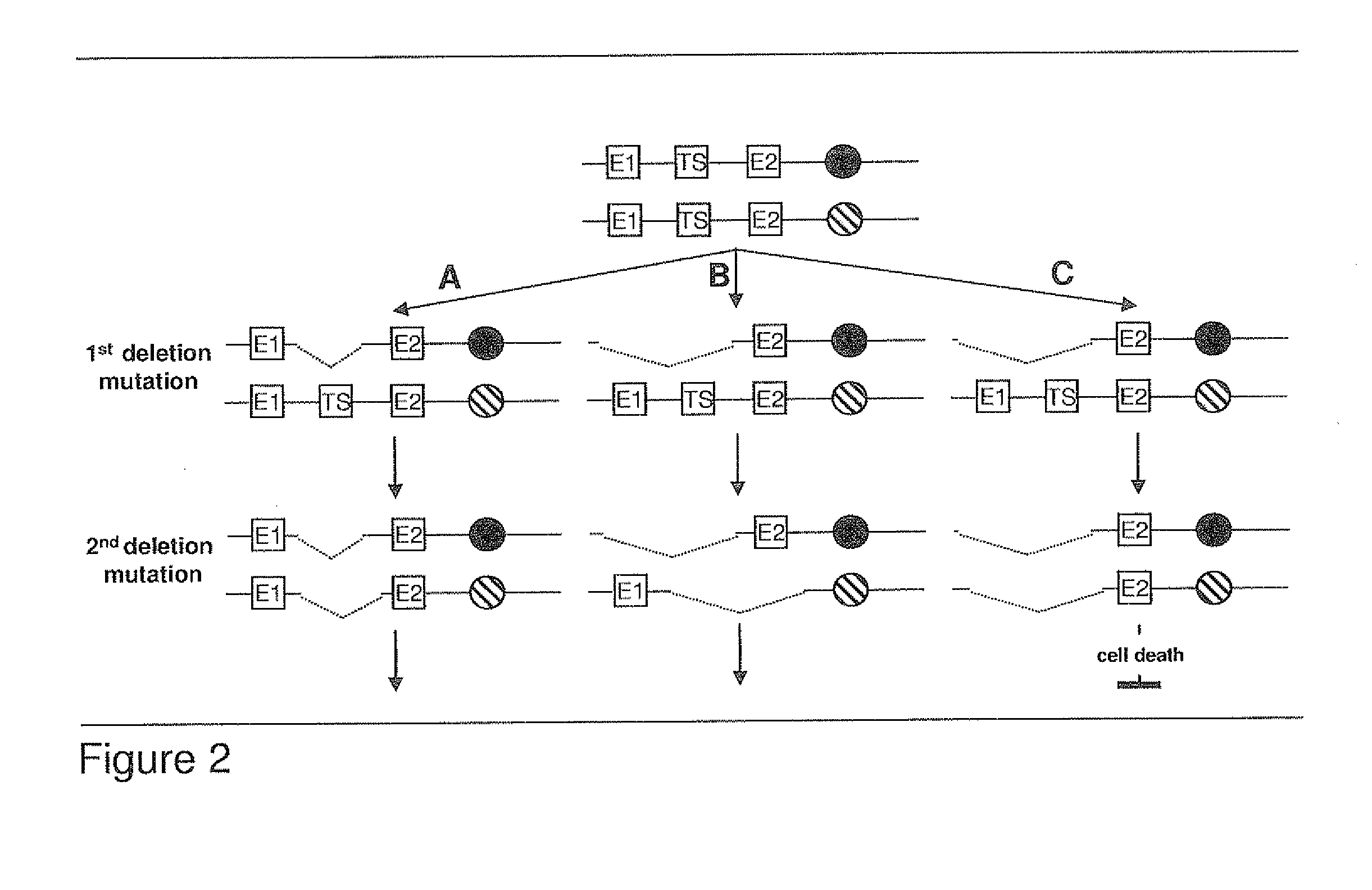
Most clinically distinguishable malignant tumors are characterized by specific mutations, specific patterns of chromosomal rearrangements and a predominant mechanism of genetic instability. It has been suggested that the internal dynamics of genomic modifications as opposed to the external evolutionary forces have a significant and complex impact on Darwinian species evolution. A similar situation can be expected for somatic cancer evolution as the key mechanisms encountered in species evolution such as duplications, rearrangements or deletions of genes also constitute prevalent mutation mechanisms in cancers with chromosomal instability. The invention is an algorithm which is based on a systems concept describing the putative constraints of the cancer genome architecture on somatic cancer evolution. The algorithm allows the identification of therapeutic target genes in individual cancer patients which do not represent oncogenes or tumor suppressor genes but have become putative therapeutic targets due to constraints of the cancer genome architecture on individual somatic cancer evolution. Target genes or regulatory elements may be identified by their designation as essential genes or regulatory elements in cancer cells of the patient but not in normal tissue cells or they may be identified by their impact on the process of somatic cancer evolution in individual patients based on phylogenetic trees of somatic cancer evolution and on the constructed multilayered cancer genome maps. The algorithm can be used for delivering personalized cancer therapy as well as for the industrial identification of novel anti-cancer drugs. The algorithm is essential for designing software programs which allow the prediction of the natural history of cancer disease in individual patients.
Owner:RUBBEN ALBERT
Method and kit for detecting chromosome rearrangement
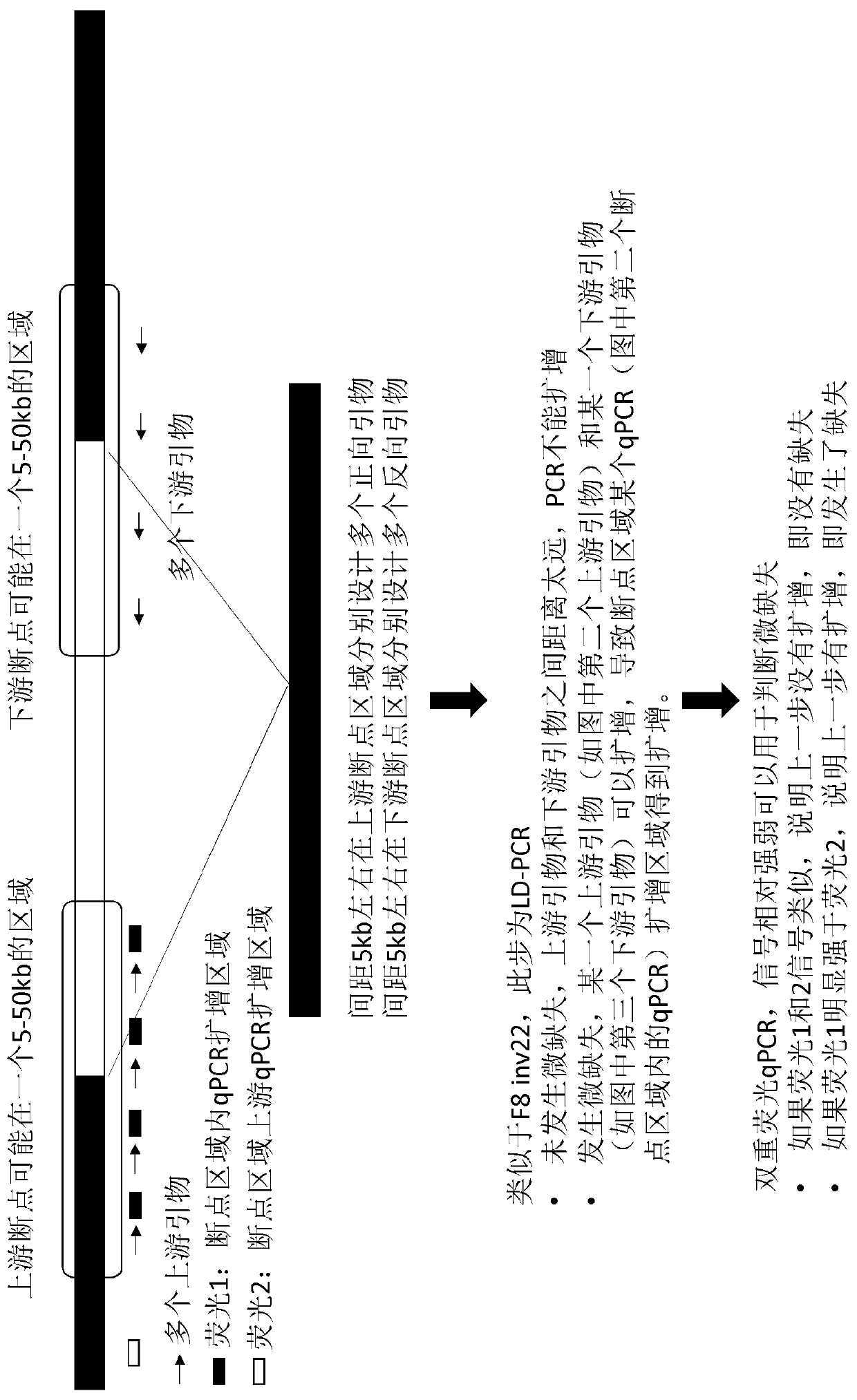
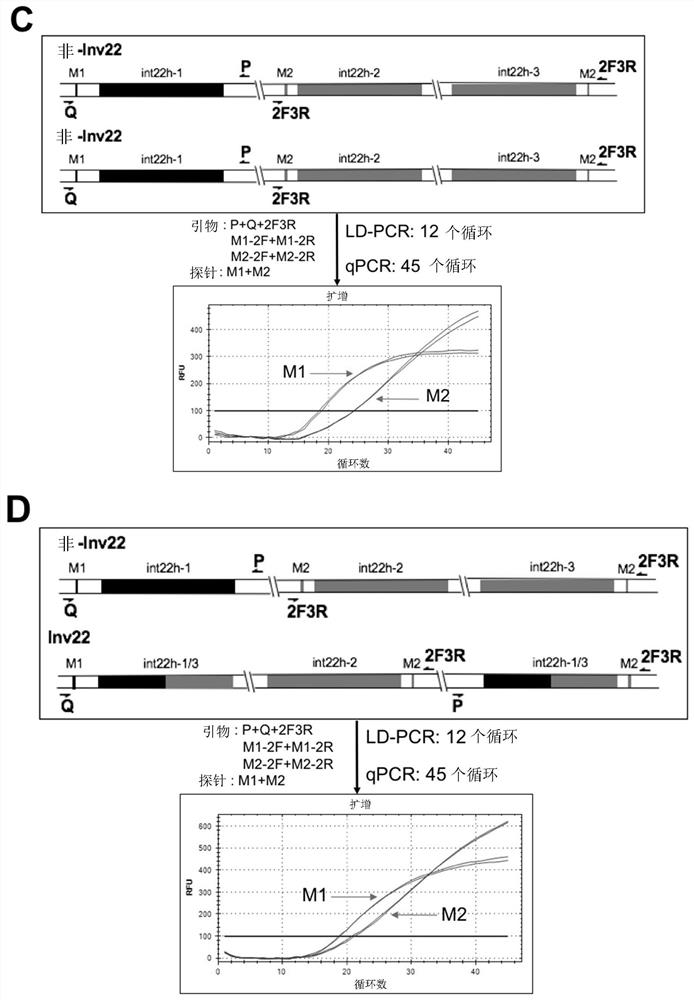
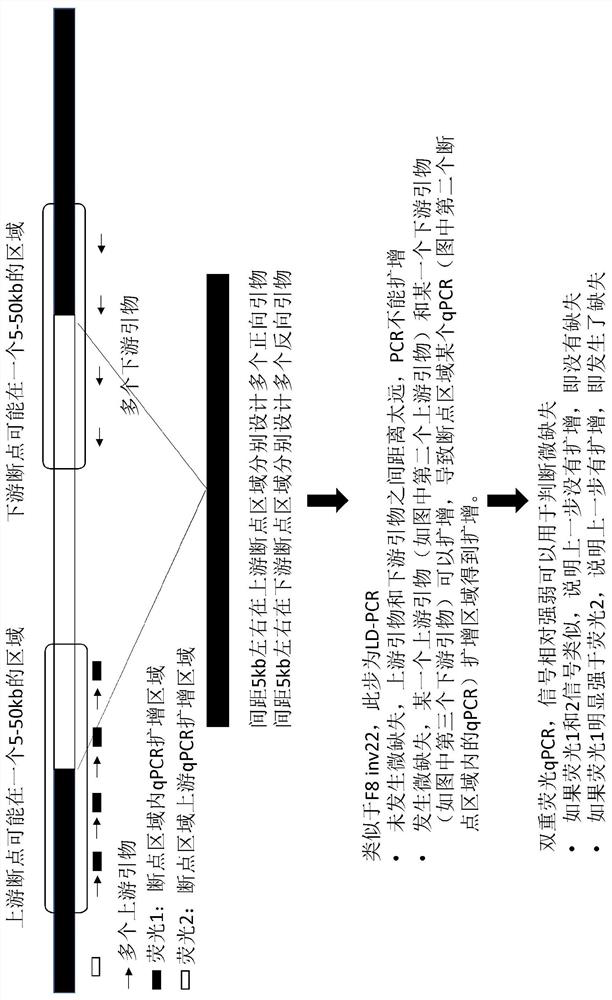
ActiveCN111020022ASave on LD-PCR processImprove throughputMicrobiological testing/measurementDNAChromosomal rearrangement
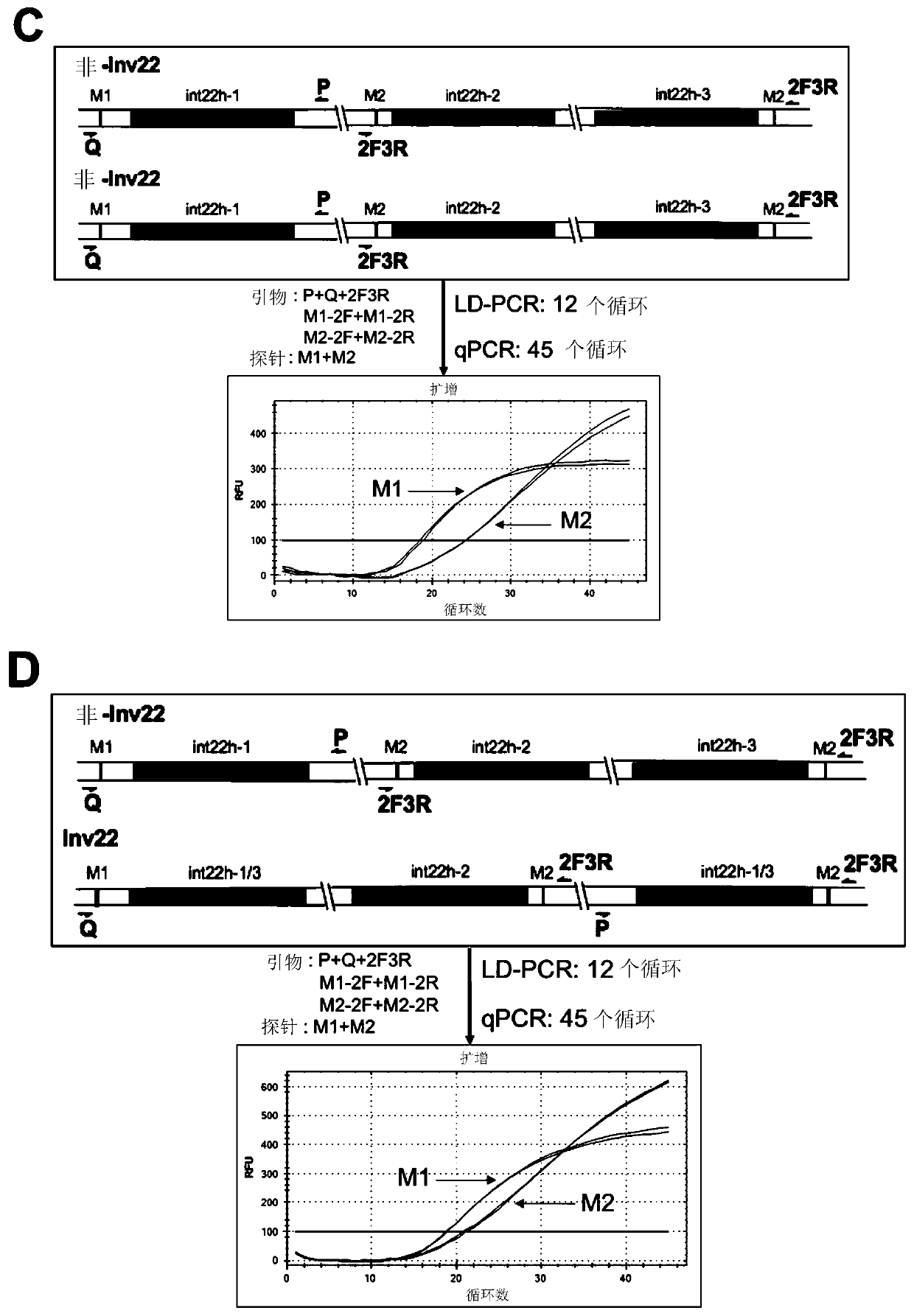
The invention relates to a method and a kit for detecting chromosome rearrangement, specifically to a method for detecting chromosome rearrangement. The method comprises the following steps: obtainingsample DNA from a sample from a subject; carrying out long-distance PCR (LD-PCR) and quantitative PCR (qPCR) on the DNA of the sample; and comparing the detection result with the result of a normal control sample, and deducing whether chromosomal rearrangement is present in the sample DNA according to the result of the comparison, wherein the LD-PCR and the qPCR can be carried out successively orin the same reaction container. The invention also relates to a kit, a primer and a probe for carrying out the method.
Owner:WENZHOU MEDICAL UNIV
System and method for secondary analysis of nucleotide sequencing data
ActiveUS20190385699A1Microbiological testing/measurementBiostatisticsReference genome sequenceTime efficient
Disclosed herein are systems and methods for performing secondary analyses of nucleotide sequencing data in a time-efficient manner. Some embodiments include performing a secondary analysis iteratively while sequence reads are generated by a sequencing system. Secondary analyses can encompass both alignment of sequence reads to a reference sequence (e.g., the human reference genome sequence) and utilization of this alignment to detect differences between a sample and the reference. Secondary analysis can enable detection of genetic differences, variant detection and genotyping, identification of single nucleotide polymorphisms (SNPs), small insertions and deletion (indels) and structural changes in the DNA, such as copy number variants (CNVs) and chromosomal rearrangements.
Owner:ILLUMINA INC
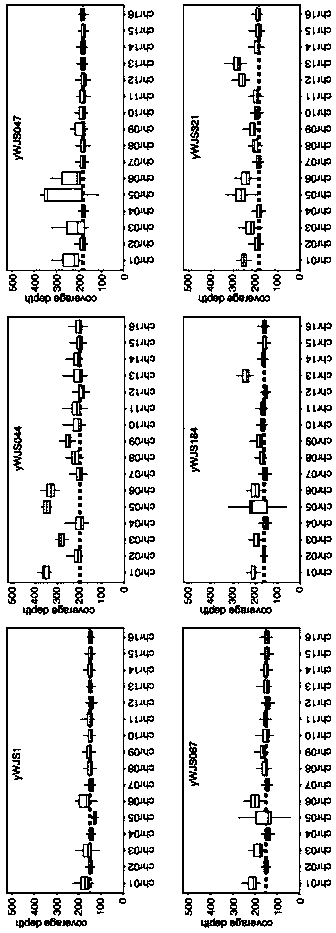
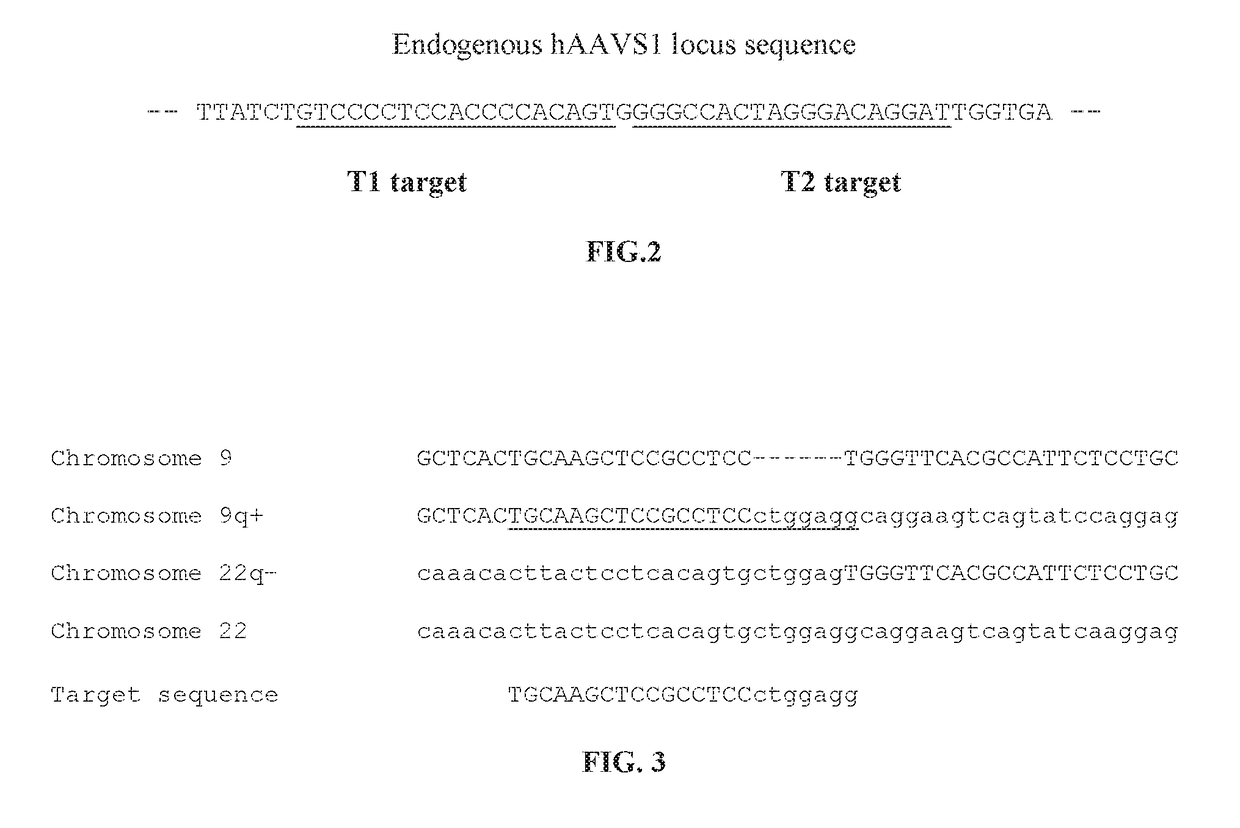
Methods and compositions for selectively eliminating cells of interest
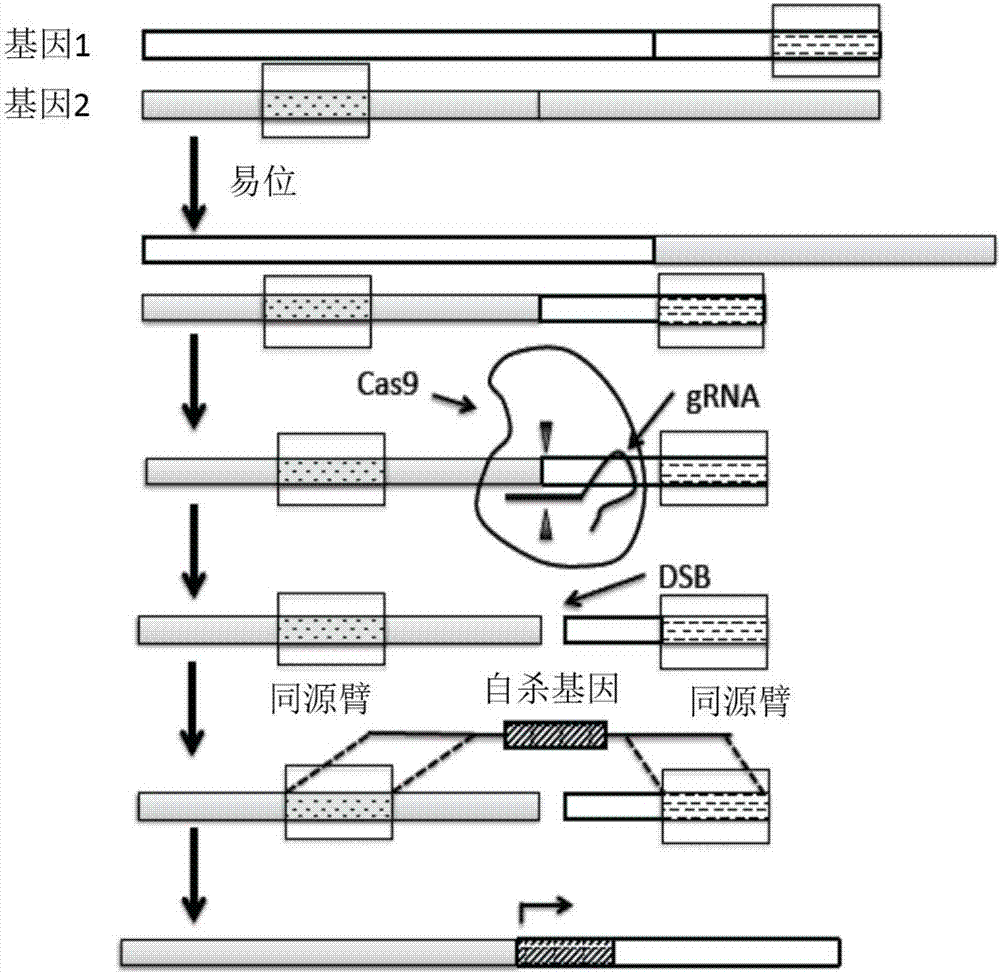
The present disclosure provides novel compositions and methods suitable for specifically eliminating target cells (e.g., cancer cells) without affecting non-target cells (e.g., non-cancer cells). For example, CRISPR system and the compositions of the present disclosure can be employed to specifically introduce a suicidal gene into a cancer cell in the loci of a cancer-specific target sequence, which as a result of chromosomal re-arrangement or translocation in a cancer cell presents a cancer specific sequence for a guide RNA and CAS to be recognized and such sequence is absent in a non-cancer cell. Consequently, the specific introduction of the composition(s) to cancer-specific site(s) and integration of suicide gene in the target genome, which is inapplicable to normal cells for lack of the site(s), leads to selective elimination of cancer cells but not non-cancer cells, and therefore render novel therapeutic methods and compositions for cancer treatment.
Owner:朱坚
Hotspots for chromosomal rearrangement in breast and ovarian cancers
InactiveUS20190345562A1Minimizing false positive resultMaximize chanceMicrobiological testing/measurementBiostatisticsSpecific chromosomeCancer genome
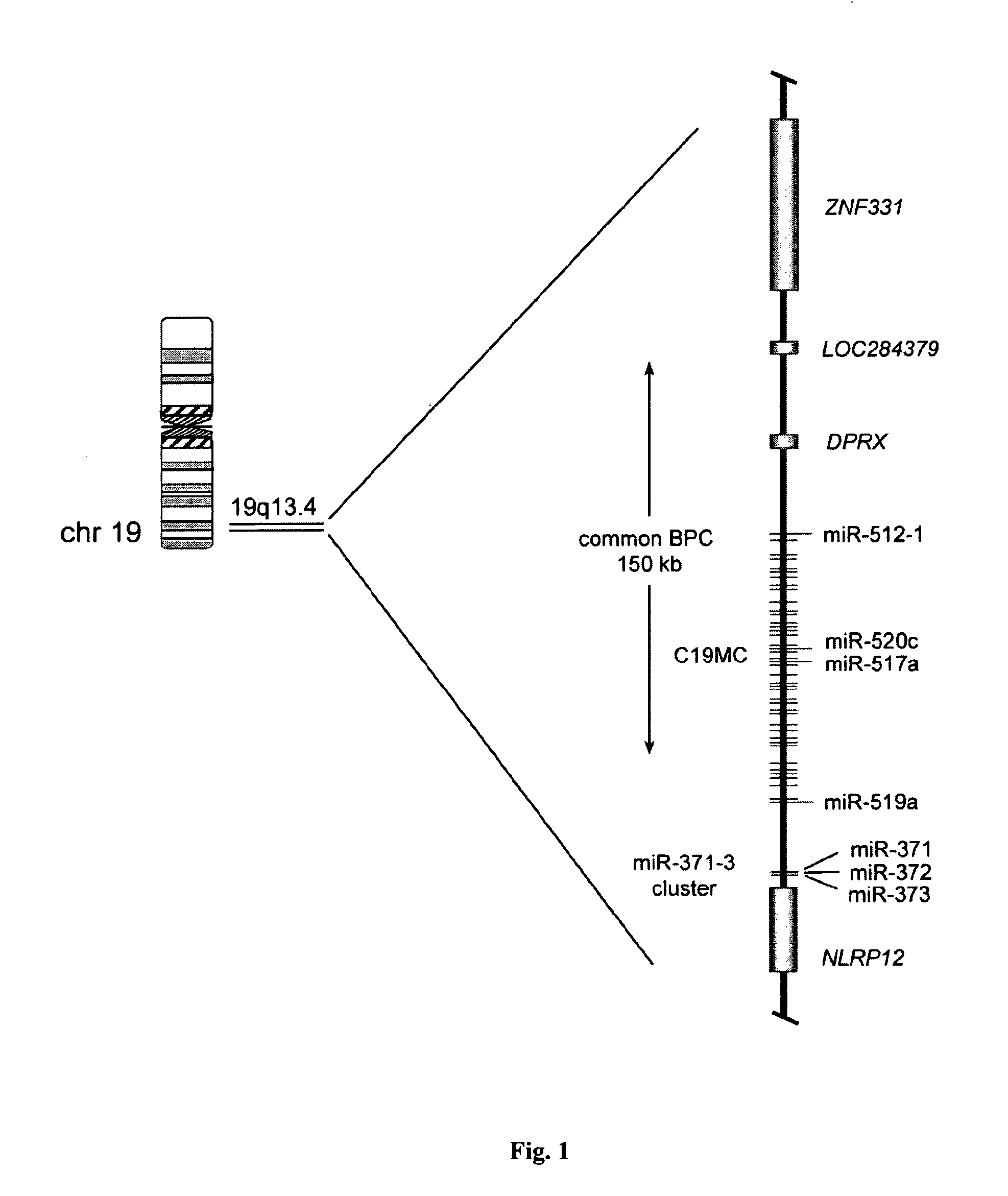
The invention relates to the classification of breast and ovarian tumours, and in particular to the use of particular rearrangement signatures to identify tumours as deficient in homologous recombination repair (HR-deficient). The inventors have identified particular chromosomal “hotspots” of recombination in breast and ovarian cancers which permit the homologous recombination repair status of a cancer to be assessed by determining the presence of recombination events within those specific hotspots, rather than by analysing the entire cancer genome for the presence of rearrangement signatures as a whole.
Owner:GENOME RES LTD
Methods and kits for detecting chromosomal rearrangements
ActiveCN111020022BIncreased sensitivityReduce the risk of contaminationMicrobiological testing/measurementDNAChromosomal rearrangement
Provided in the present invention are a method and kit for detecting chromosome rearrangement. Specifically, the method for detecting chromosomal rearrangement of the present invention comprises: obtaining sample DNA from a sample of a subject; performing long-distance PCR (LD-PCR) and quantitative PCR (qPCR) on the sample DNA; comparing a detection result with a result of a normal control sample; and from the result of the comparison, inferring whether a chromosomal rearrangement is present in the sample DNA. The LD-PCR and qPCR may be performed in succession or in the same reaction container. Further provided in the present invention are a kit for performing the method, as well as primers and probes.
Owner:WENZHOU MEDICAL UNIV
Methods and compositions for enrichment of amplification products
In some aspects, the present disclosure provides methods for enriching amplicons, or amplification products, comprising a concatemer of at least two or more copies of a target polynucleotide. In some embodiments, a method comprises sequencing the amplicons comprising at least two or more copies of a target polynucleotide. In some embodiments, the target polynucleotides comprise sequences resulting from chromosome rearrangement, including but not limited to point mutations, single nucleotide polymorphisms, insertions, deletions, and translocations including fusion genes. In some aspects, the present disclosure provides compositions and reaction mixtures useful in the described methods.
Owner:ACCURAGEN HLDG LTD
DNA microarray-based identification and localization of balanced translocation breakpoints
Owner:UNIV OF WASHINGTON
Microrna-based methods and compositions for the diagnosis, prognosis and treatment of tumor involving chromosomal rearrangements
ActiveUS20120295949A1Organic active ingredientsMicrobiological testing/measurementAbnormal tissue growthMedicine
Provided are novel methods and compositions for the diagnosis, prognosis and treatment of tumor involving a chromosomal rearrangement, in particular a tumor or neoplasia of the thyroid gland. In addition, methods of identifying anti-tumor agents are described.
Owner:UNIV OF BREMEN
An optimized method for efficiently and rapidly constructing a library of homogeneous mutants with high saturation and high resistance in stepgrass
ActiveCN105123530BEasy accessLarge capacityPlant tissue cultureHorticulture methodsEthylmethane SulfonateHigh resistance
The invention discloses an optimization method for efficiently and quickly building an ophiopogon japonicus saturation high resistance homogeneous mutant library. Ethylmethane sulfonate (EMS) and <60> Co-gamma ray are physical and chemical mutagenic agents which are most widely used, efficient and stable. The EMS can generate a plurality of point mutations on one genome, the generated mutations can be randomly distributed on the whole genome, the distribution is even, the density is high, the cost is low, and the obtaining of allelic mutants is facilitated, especially, a saturated mutant library can be obtained by a smaller mutant population; Gamma ray mutagenesis mainly causes the loss of DNA fragments and chromosomal rearrangement and is specially suitable for the building of a mutant library of which multiple family gene functions are lost; therefore, by using the two method simultaneously to make their respective advantages complementary to each other, the capacity of the mutant library is increased, the demands of the saturated mutant library are satisfied, the content of the saturated mutant library is enriched, and moreover, the obtained mutants do not involve a GM event, so the safety is high and the application is convenient.
Owner:JIANGSU POLYTECHNIC COLLEGE OF AGRI & FORESTRY
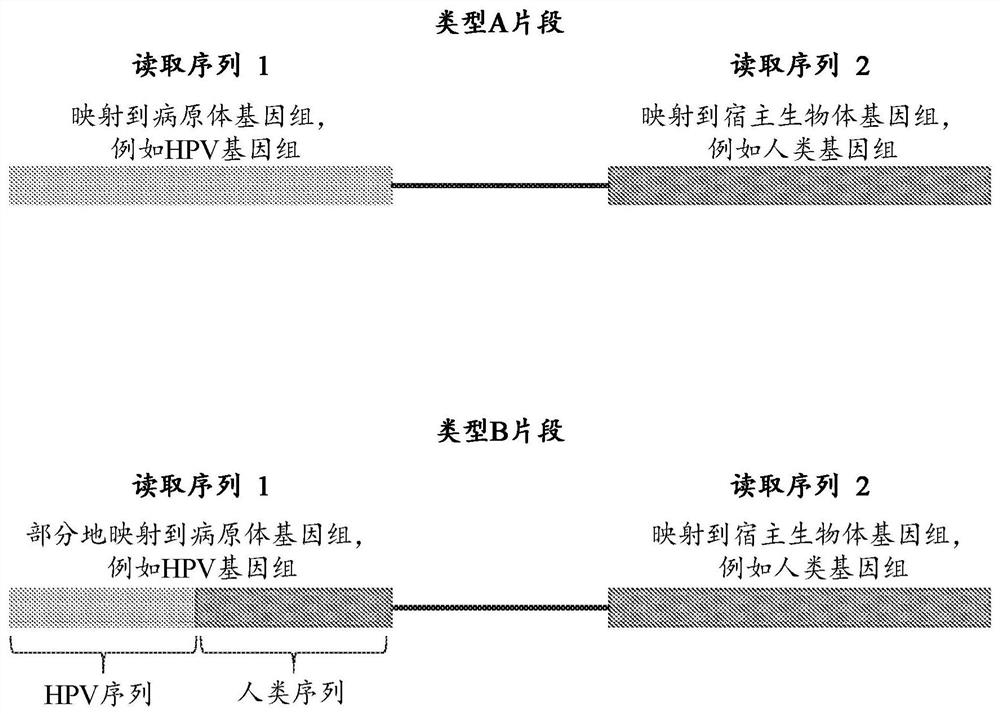
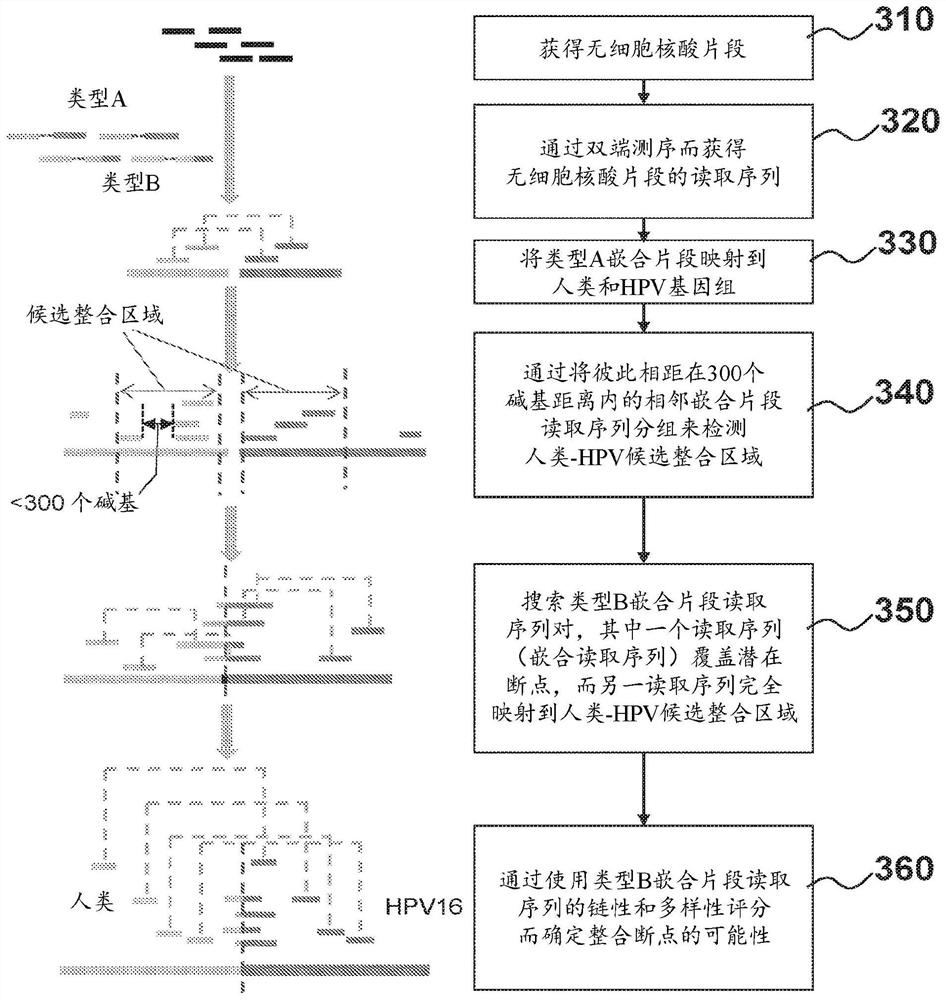
Nucleic acid rearrangement and integration analysis
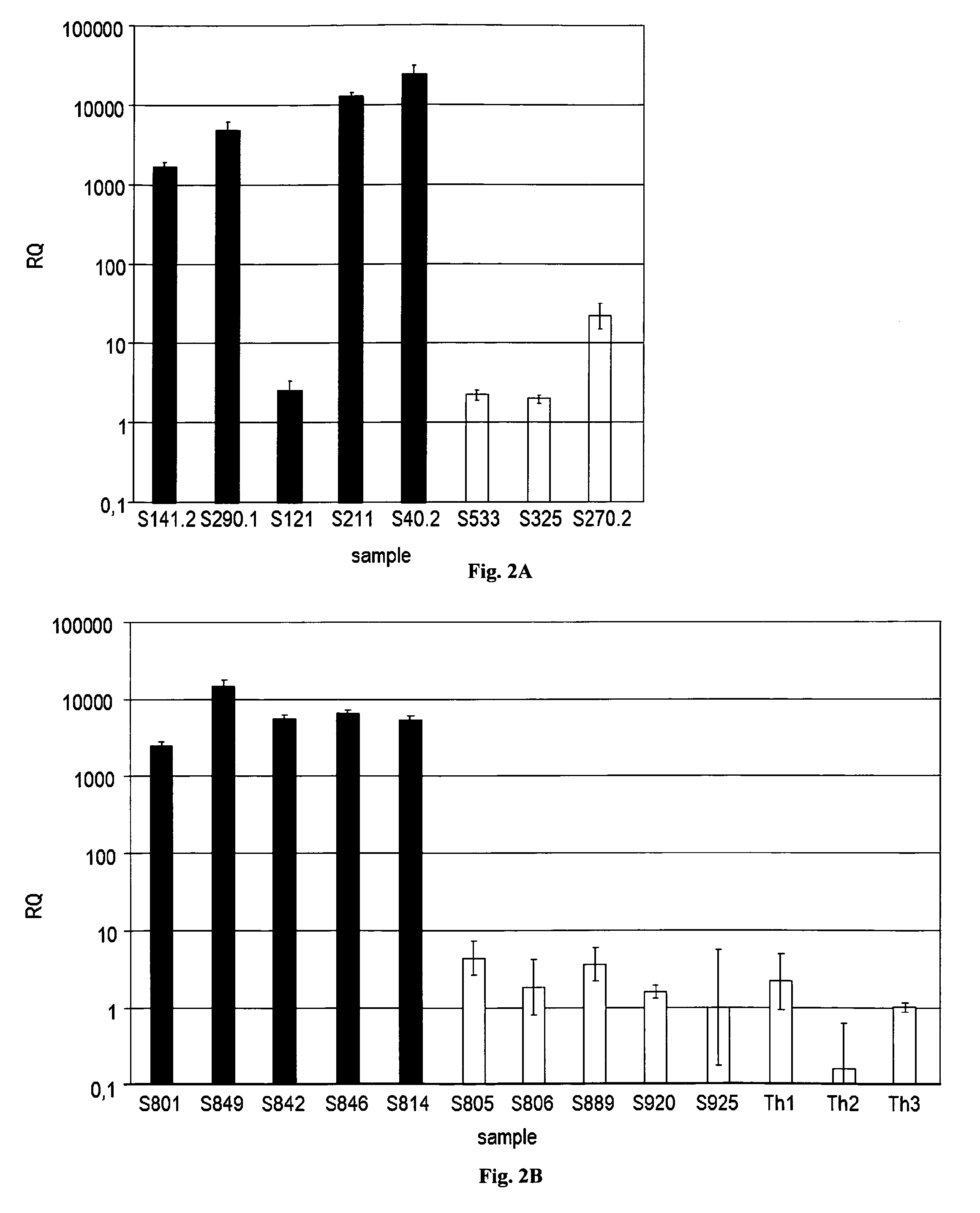
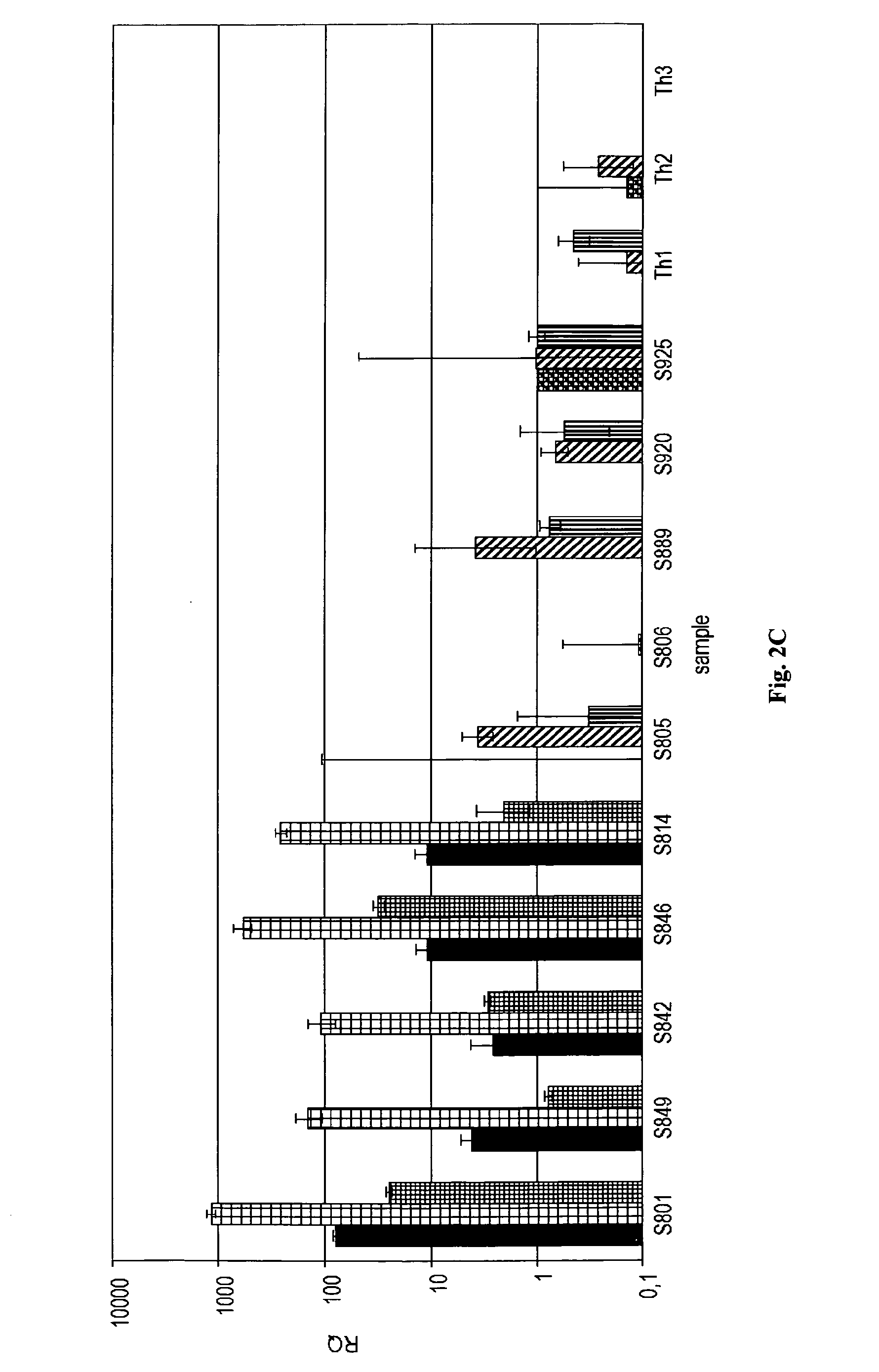
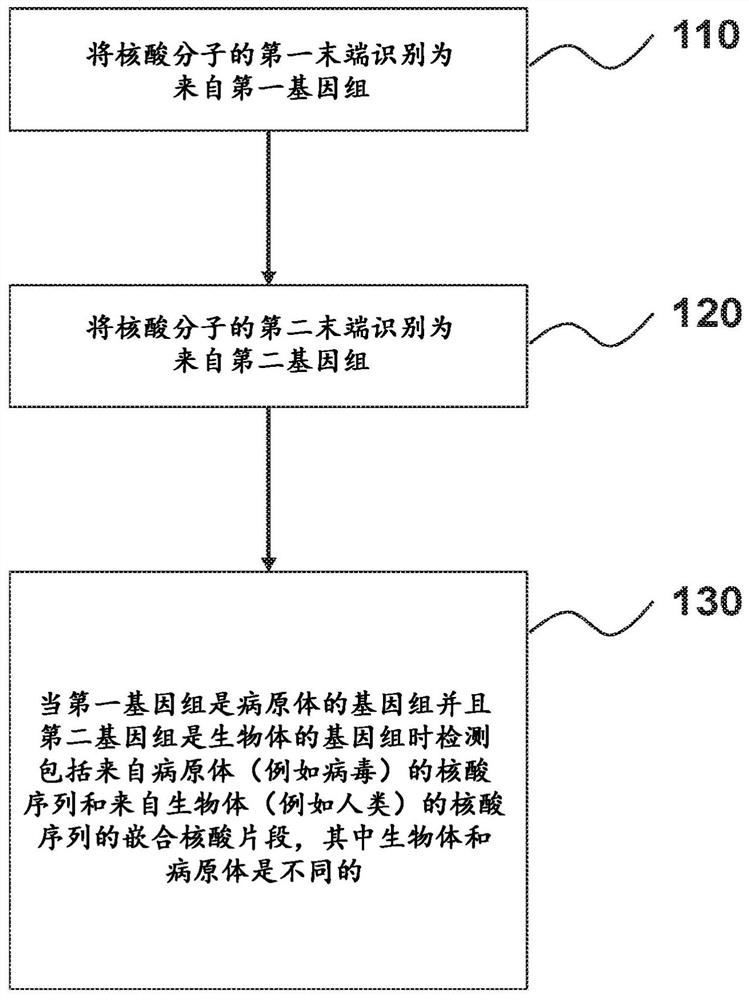
Provided herein are methods and systems for identifying chimeric nucleic acid fragments, e.g., organism-pathogen chimeric nucleic acid fragments and chromosomal rearrangement chimeric nucleic acid fragments. Also provided herein are methods and systems relating to determining a pathogen integration profile or a chromosomal rearrangement in a biological sample and determining a classification of pathology based at least in part on a pathogen integration profile or a chromosomal rearrangement in a biological sample. In certain aspects of the present disclosure, cell-free nucleic acid molecules from a biological sample are analyzed.
Owner:GRAIL INC
Gene element and application of same to variation of chromosome number, copy number and structure of Saccharomyces cerevisiae
ActiveCN108913611ARealize the structureAchieving Continuous VariationFungiMicroorganism based processesBiotechnologyYeast chromosome
The invention especially relates to a generation method for the variation of the chromosome number of Saccharomyces cerevisiae, belonging to the field of synthetic biology. According to the invention,in virtue of the characteristics of genome rearrangement of Saccharomyces cerevisiae and through genome rearrangement of the ring chromosomes of Saccharomyces cerevisiae, an aneuploid Saccharomyces cerevisiae strain with chromosome number variation is obtained, the continuous variation of the circular chromosome copy number of Saccharomyces cerevisiae is realized, a Saccharomyces cerevisiae strain with continuously evolved phenotypes is obtained, and correlation between structural variation and phenotype enhancement is revealed. According to the invention, the variation of the chromosome number, the copy number and the structure of Saccharomyces cerevisiae is realized and the structural variation is revealed by using a chromosome rearrangement method, and a novel research means is provided for obtaining aneuploid Saccharomyces cerevisiae strains and studying the relationship between chromosome structure variation and functions.
Owner:TIANJIN UNIV
Methods and compositions for selectively eliminating cells of interest
The present disclosure provides novel compositions and methods suitable for specifically eliminating target cells (e.g., cancer cells) without affecting non-target cells (e.g., non-cancer cells). For example, CR-ISPR system and the compositions of the present disclosure can be employed to specifically introduce a suicidal gene into a cancer cell in the loci of a cancer-specific target sequence, which as a result of chromosomal re-arrangement or translocation in a cancer cell presents a cancer specific sequence for a guide RNA and CAS to be recognized and such sequence is absent in a non-cancer cell. Consequently, the specific introduction of the composition(s) to cancer-specific site(s) and integration of suicide gene in the target genome, which is inapplicable to normal cells for lack of the site(s), leads to selective elimination of cancer cells but not non-cancer cells, and therefore render novel therapeutic methods and compositions for cancer treatment.
Owner:ZHU JAMES
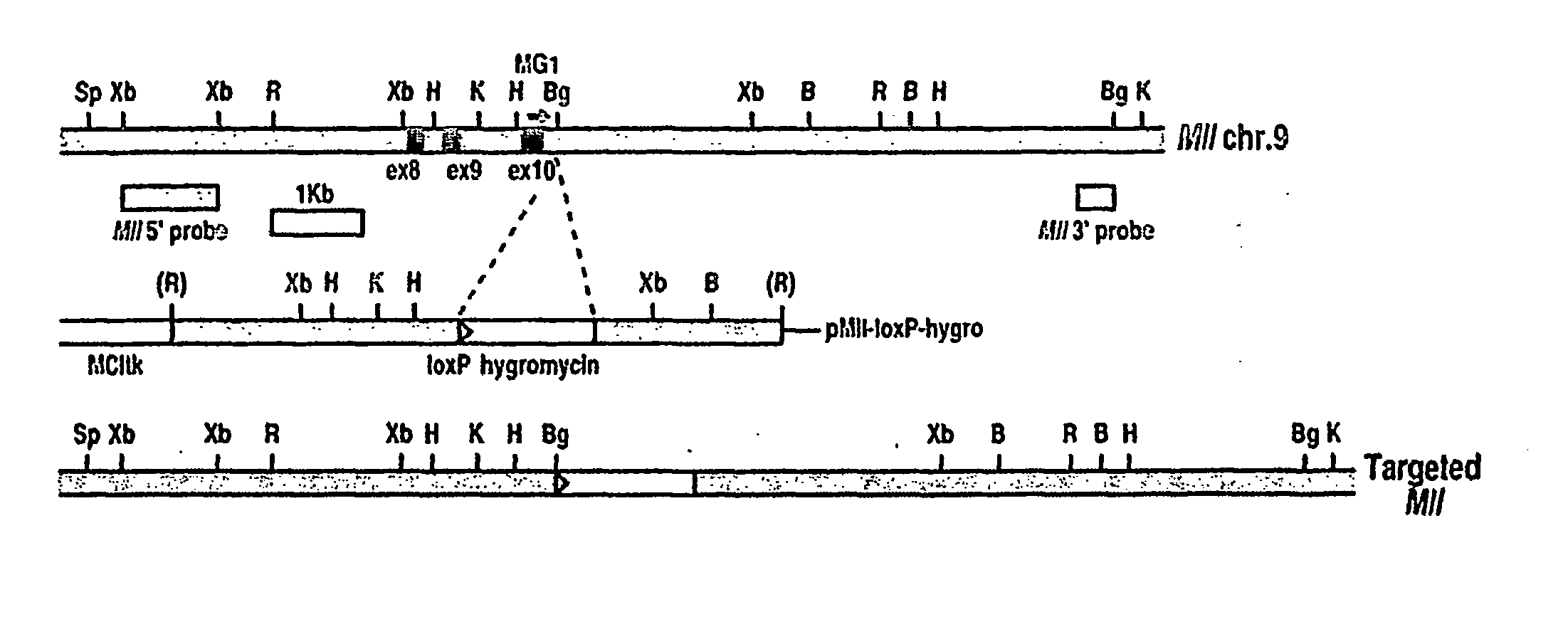
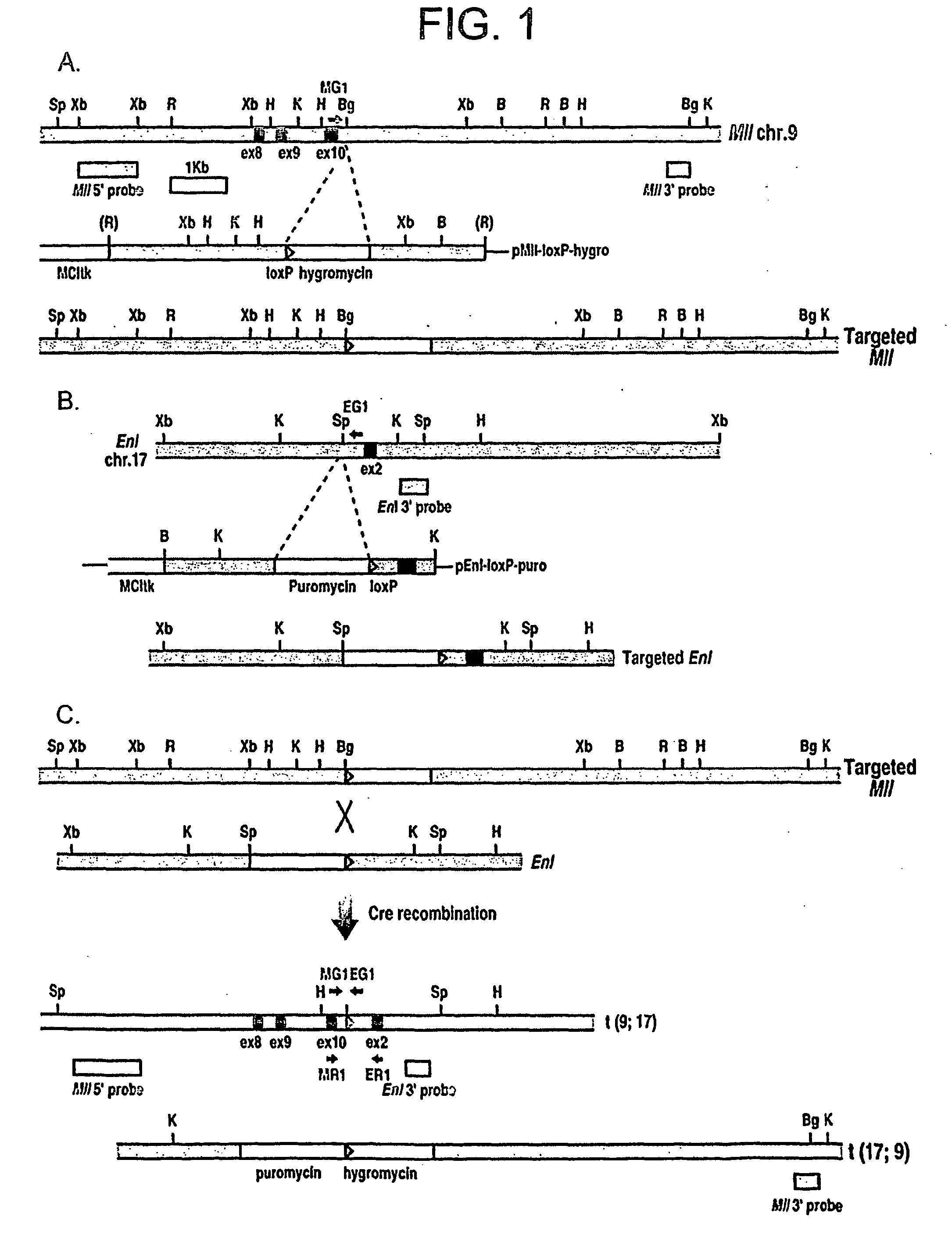
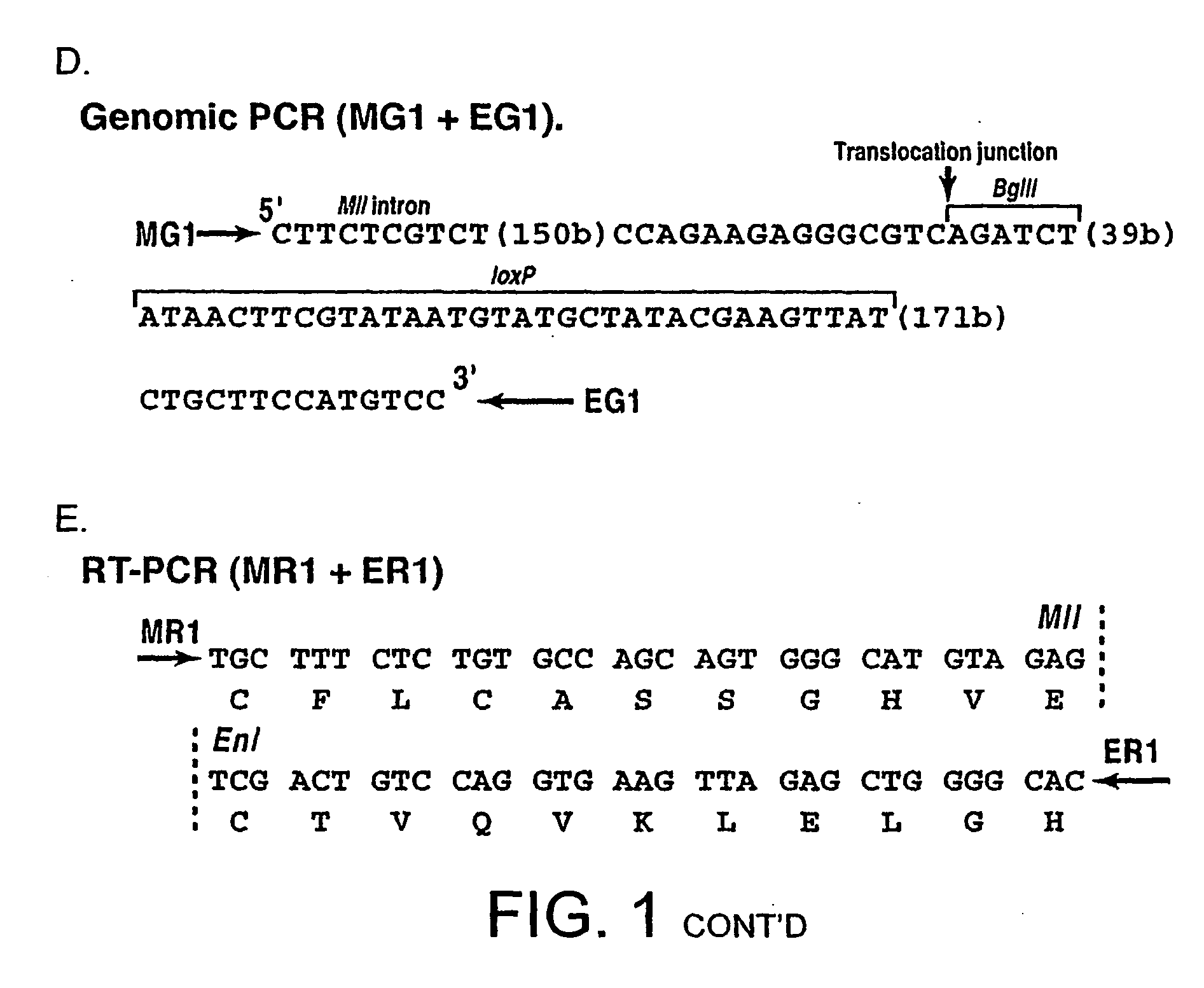
Tumor model with chromosomal rearrangement and uses thereof
InactiveUS20060179495A1Rapid onset and high penetrance of leukaemogenesisRapid and reliable resultNucleic acid vectorAnimal husbandryAbnormal tissue growthHuman animal
The invention relates to a method for generating a non-human animal model of a chromosomal rearrangement, comprising creating a transgenic non-human mammal expressing a site-specific recombinase under the control of a cell type specific promoter, and having sites recognized by the recombinase present in its genome such that a chromosomal rearrangement is catalysed by the recombinase.
Owner:MEDICAL RESEARCH COUNCIL
Treatment of cancers characterized by chromosomal rearrangement of the NUT gene
Owner:DANA FARBER CANCER INST INC +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com