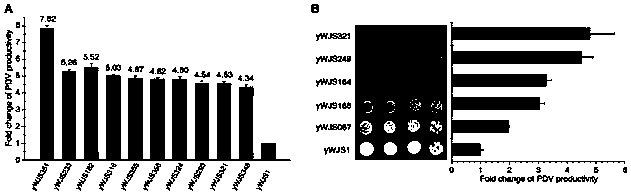Gene element and application of same to variation of chromosome number, copy number and structure of Saccharomyces cerevisiae
A genetic element, technology of Saccharomyces cerevisiae, applied in the field of synthetic biology, can solve the problem of high lethality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1 Haploid Saccharomyces cerevisiae phenotype continuous evolution method
[0054] The invention provides a research method for accelerating the phenotypic evolution of strains by using synthetic Saccharomyces cerevisiae circular chromosomes, using PDV, a downstream metabolite of aromatic amino acids, as a marker to generate a large number of phenotypic variant yeast strains by inducing circular chromosome rearrangement.
[0055] 1. design primers PCR amplify the open reading frame (ORF) of vioA, vioB and vioE from the biological building block (BioBrick) component library, agarose gel reclaims DNA fragment.
[0056] 2. Design primers to amplify the promoter and terminator from Saccharomyces cerevisiae BY4741 genomic DNA, wherein there is an overlapping region of 40 base pairs (bp) between the promoter, open reading frame and terminator. DNA fragments were recovered from agarose gel.
[0057] 3. The pRS416 plasmid was digested with EcoRI restriction endonucle...
Embodiment 2
[0079] Example 2 Saccharomyces cerevisiae violacein precursor metabolism strain continuous improvement
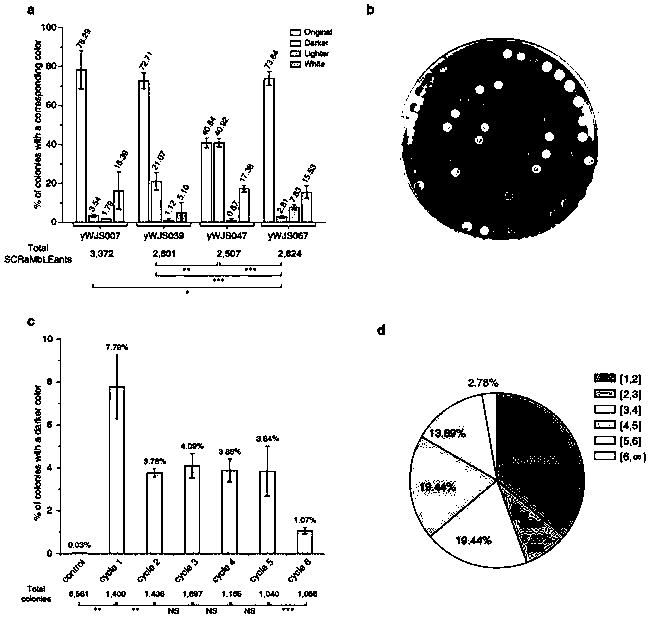
[0080] Circular synthetic yeast chromosomal rearrangements can achieve sustained improvements in the phenotype of yeast strains. We performed 5 rounds of rearrangement experiments on the strain (yWJS067), expecting to obtain a strain with improved PDV production. For the finally selected strains, the percentages of dark colonies were 7.79%, 3.76%, 4.09%, 3.88%, 3.84% and 1.07%, respectively. For each round of rearrangement experiments, the strain with the darkest visible color and similar growth fitness compared to the parental strain was selected for quantification of PDV productivity by HPLC. After six rounds of rearrangement experiments, we obtained a strain with about seven-fold increase in PDV production, and the strain had a good growth phenotype.
[0081] Table 4 figure 2 aRaw data
[0082]
[0083] table 5 figure 2 bRaw data
[0084]
[0085] Ring synthe...
Embodiment 3
[0086] Example 3 Saccharomyces cerevisiae chromosome number continuous variation
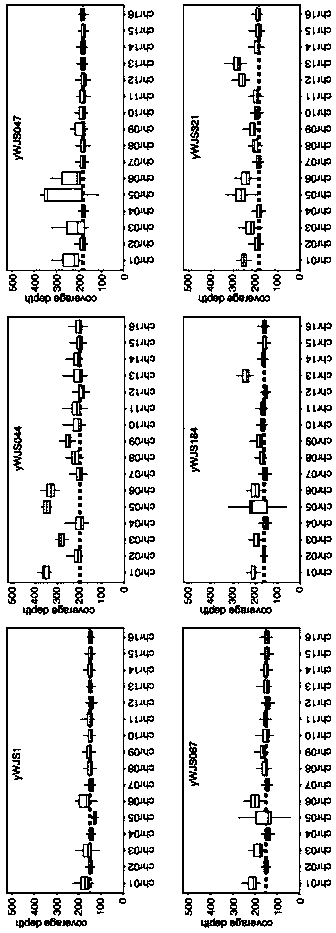
[0087]In order to reveal the genetic variation generated in ring_synV in depth, the applicant conducted whole genome sequencing (WGS) analysis on some induced rearrangement strains (SCRaMbLEants), yWJS044, yWJS047, yWJS067, yWJS184 and yWJS321. Among the rearranged strains analyzed, yWJS044, yWJS047 and yWJS067 were obtained from the starting strain yWJS1 after the first round of SCRaMbLE, yWJS184 was obtained from yWJS067 after the third round of SCRaMbLE, and yWJS321 was obtained from yWJS184 , the rearrangement strain obtained after the fifth round of SCRaMbLE.
[0088] image 3 The middle is a boxplot based on the WGS results, showing the coverage depth of different chromosomes, representing the copy number of different chromosomes, showing that the haploid S. cerevisiae synthetic ring V chromosome SCRaMbLE will lead to chromosome aneuploidy. Strains yWJS044 (aneuploid chromosomes: I, III,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com