Method for identifying industrially cultivated strains of hypsizygus marmoreus by using SSR molecular marker
A technology of shimeji mushrooms and strains, which is applied in the field of molecular biology identification of strains to achieve the effect of protecting intellectual property rights and enhancing competitiveness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
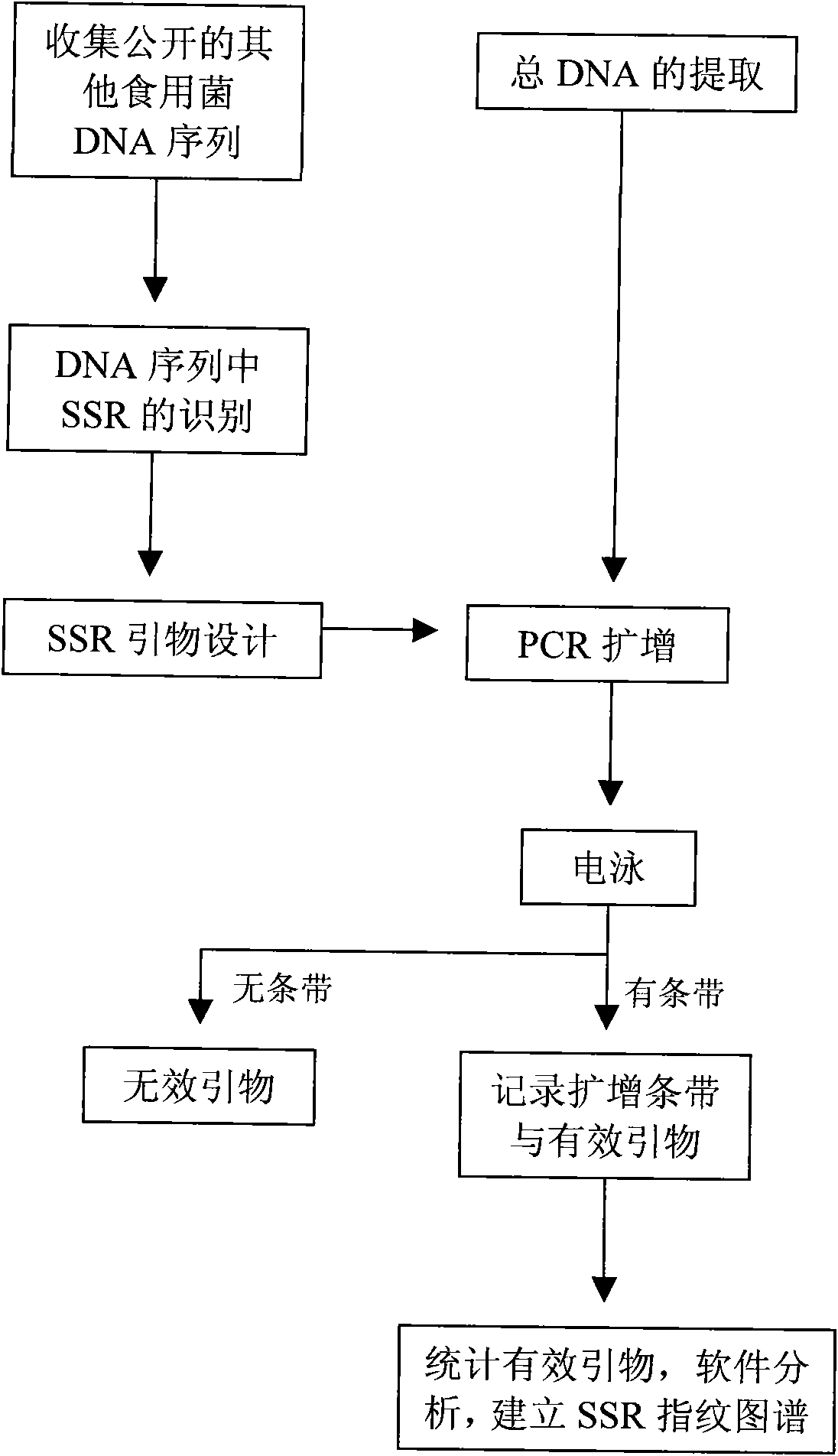
[0049] 1. DNA sequence data collection of similar species
[0050] Downloaded from the Fungal Genome Project website FGP (http: / / fungalgenomics.concordia.ca) and NCBI (http: / / www.ncbi.nlm.nih.gov) the length of mushrooms and other similar species of eutrophicum greater than 150bp, 5' and 3' end 50bp does not contain (T) 5 or (A) 5 DNA sequences with close relatives such as Lentinula edodes, Pleurotus otreatus, and Coprinus cinereus.
[0051] 2. Discover SSR in DNA sequence
[0052] Use the SSR identification tool SSRIT (Simple Sequence Repeat Identification Tool) provided by the GRAMENE website to perform SSR identification on the downloaded EST sequence (http: / / www.gramene.org / db / searches / ssrtool), or use SSRHunter 1.3 software combined with manual SSR find. The identification criteria were the minimum lengths of single, di, tri, quad, penta and hexanucleotide repeat motifs were 15, 8, 5, 4, 3 and 3 times, respectively.
[0053] 3. PCR primer design
[0054] Select elig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com