Method of identifying the species of plant belonging to the genus uncaria
A technology of Uncaria, plants, applied in the field of identifying species of Uncaria plants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] Material
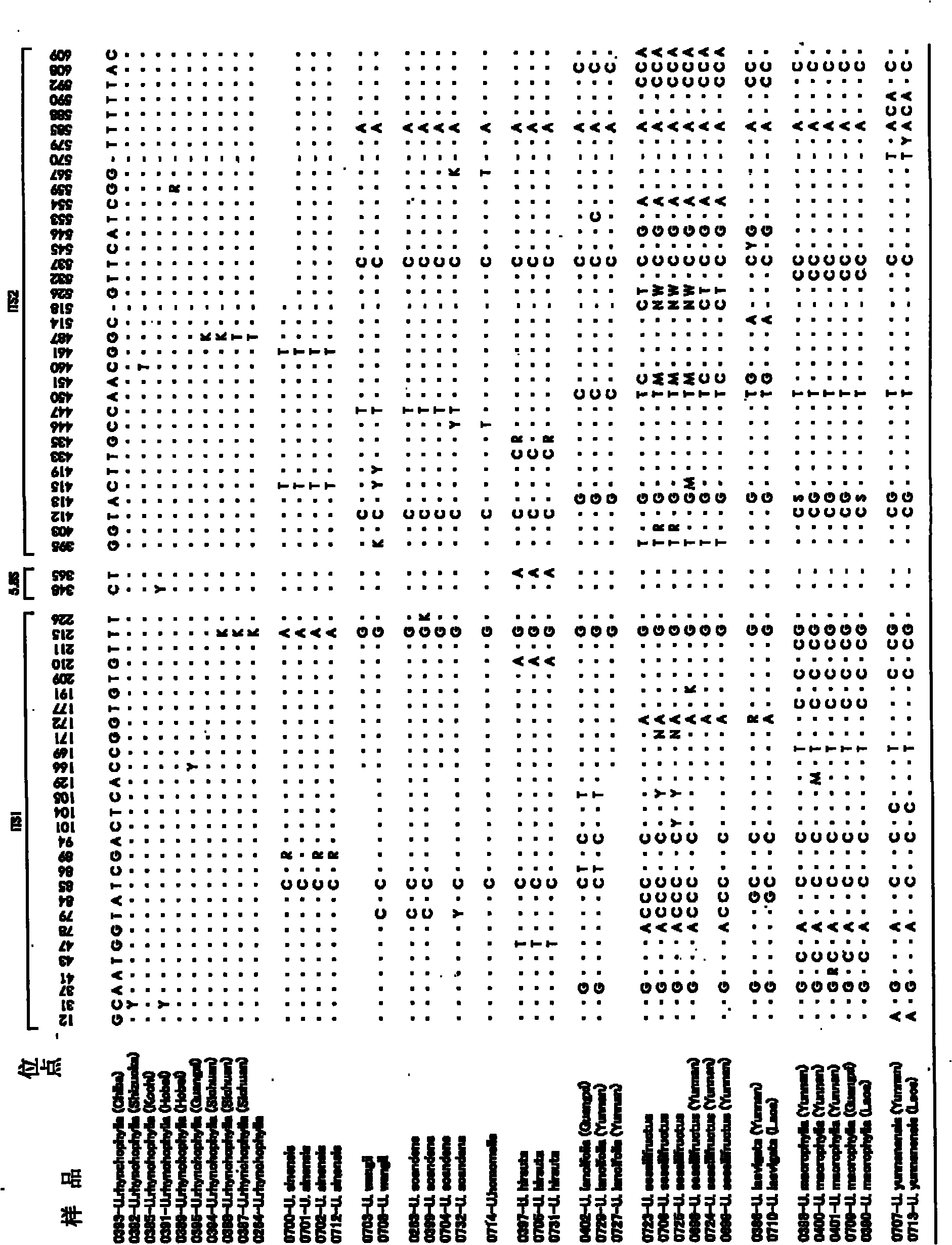
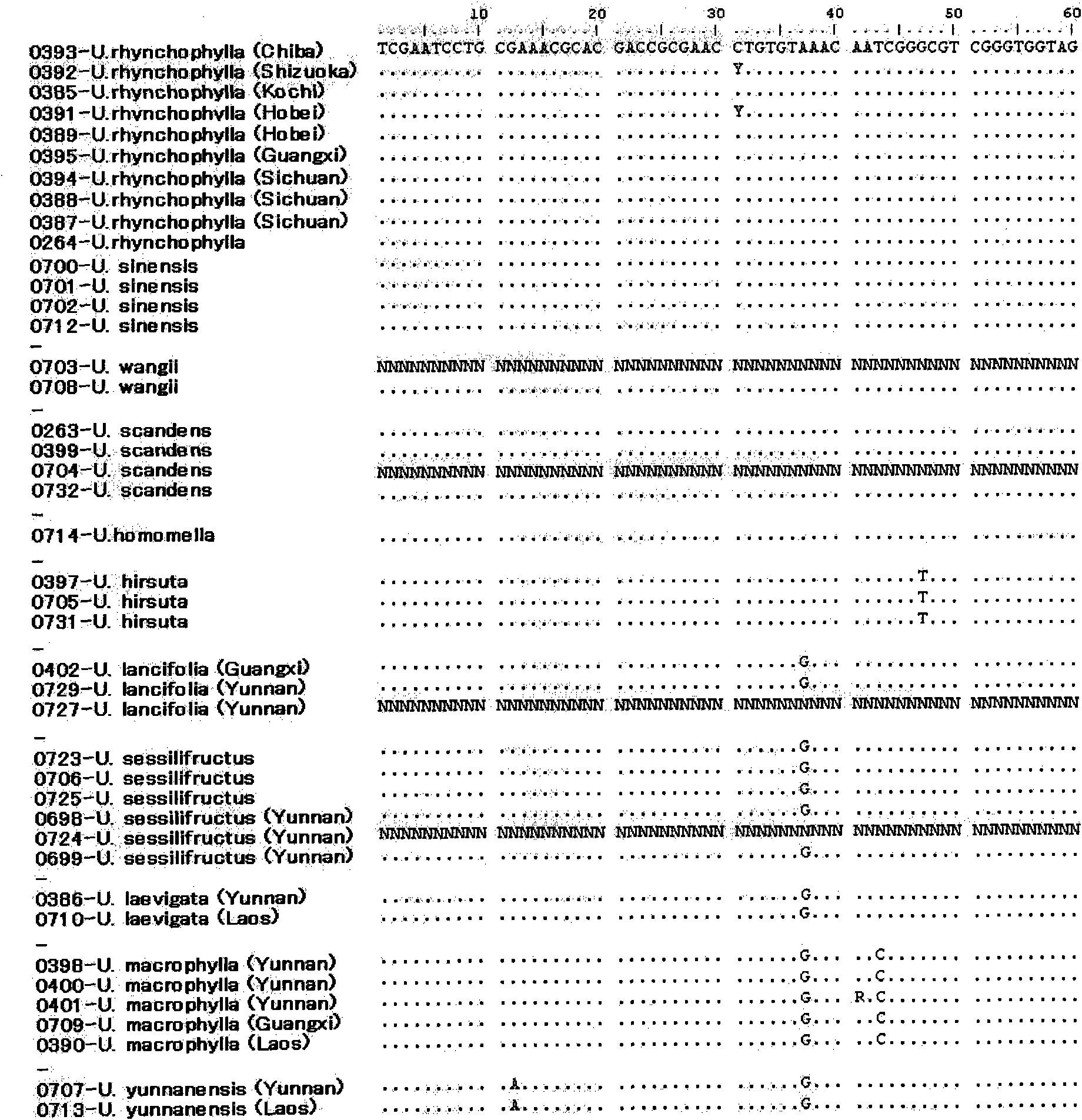
[0091] Use wax leaf specimens that can be identified according to the morphology to study the variation sites that are conducive to identification. The samples used for the study are shown in Table 3. Except for Uncaria homomella, for each species, the base sequences of the ITS region were determined for a plurality of samples, and the species-specific site was studied.
[0092] table 3
[0093]
[0094] method
[0095] About 200 mg of leaves or pericarp were collected from the samples, and the total DNA was extracted using DNeasy (registered trademark) PlantMini Kit (Qiagen). Regarding the PCR amplification of the ITS region, since the amplifiable length varies depending on the state of the sample, the amplification is performed in the following procedure. Also, for samples that are difficult to amplify, try to purify the entire DNA using the Plasmid Mini Kit (Qiagen).
[0096] (1) Using a primer set (ITS5 & ITS4), ITS1, 5.8S, and ITS2 were ampli...
Embodiment 2
[0116] By analyzing the nucleotide sequences of the ITS region of Diaoyinghua (crude drug) obtained from Huangjing Township, Gulin County, Sichuan Province, and Diaoyinghua (crude drug) obtained from a production company in Sichuan Province, it was identified which Uncaria plant was included.
[0117] Material
[0118] As samples, 2 samples of rattan hooks obtained from Huangjing Township, Gulin County, Sichuan Province and 2 batches of 8 samples of rattan hooks obtained from Chengdu Longquan Natural Medicine Development Co., Ltd. were used.
[0119] method
[0120] Total DNA was extracted from about 200 µg of the sample using DNeasy (registered trademark) Plant Mini Kit (Qiagen). In the PCR amplification of the ITS region, primer sets (ITS5 & ITS3) were used. The composition of the reaction solution is as follows: 5 μl of 10×Buffer, 4 μl of dNTP mix (2.5 mM), 2.5 μl×2 of primer (10 pmol / μl), 30.75 μl of 10% DMSO, 5 μl of template DNA, and 0.25 μl of Gene-Taq. The react...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com