Method for quickly amplifying target genes from genome DNA
A genome and gene technology, applied in the field of genetic engineering, can solve problems such as expensive, cumbersome PCR amplification, and low copy number
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment one: human α-glucosidase gene fragment amplification
[0019] 1. Preparation of human genomic DNA:
[0020] The blood samples of healthy people were donated by the Pediatric Laboratory of Fourth Medical University, and the whole blood genomic DNA extraction kit (solution type) was purchased from Changsha Lianbo Zhida Biotechnology Company. The genomic DNA extraction method was operated according to the kit instructions, and the obtained human genomic DNA was stored at -20°C for future use.
[0021] 2. Design of amplification primers for the 25th to 36th exon of human a-glucosidase gene:
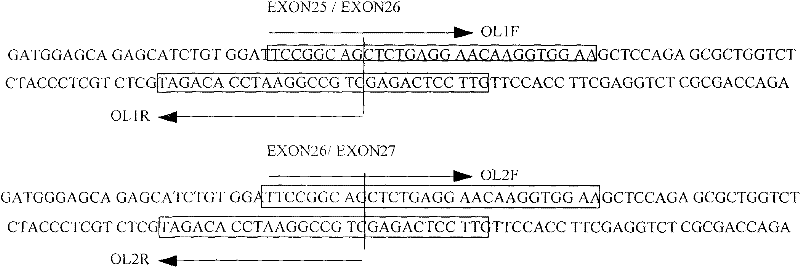
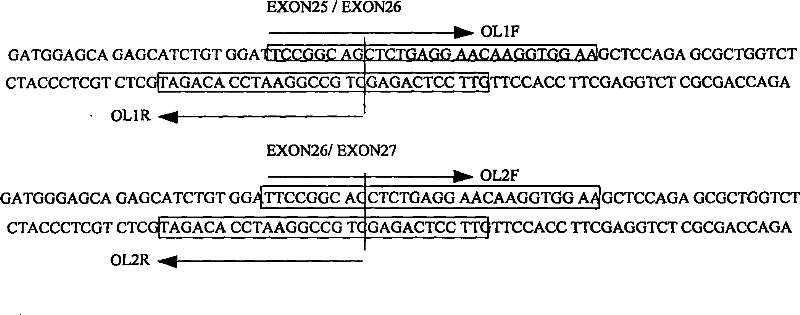
[0022] According to the exon sequence information of the found human a-glucosidase gene (NM_004668.2), overlapping primers were designed for the 12 exons respectively, and the specific operations were as follows figure 1 As shown (only schematic diagram):
[0023] The primer sequences of the 12 exons are designed as follows:
[0024] Primer name
Primer seque...
Embodiment 2
[0041] Embodiment two: the cloning of human MICA gene
[0042] 1. Preparation of human genomic DNA:
[0043] The preparation method of human genomic DNA is the same as that in Example 1.
[0044] 2. Primer design for human MICA gene cloning:
[0045] According to the found exon sequence information of human a-glucosidase gene (NM_000247.1), overlapping primers were designed for the 6 exons respectively. The design principle is as follows figure 1 shown.
[0046] The primer sequences designed for the 6 exons are as follows:
[0047] Primer name
Primer sequence
HMICA ex1F
CCCGAATTCCACTGCTTGAGCCGCTGAGA
29
HMICA ex1R
CTGTGGGGCTCAGCAGCAGCAGCTCCCGGA
27
HMICA ex2F
GCTGCTGCTGAGCCCCACAGTCTTCGTTAT
30
HMICA ex2R
GGGAATGCAAGCCTTCTTTCTGGTCCTTGA
30
HMICA ex3F
CAGAAAGAAGGCTTGCATTCCCTCCAGGAGA
31
HMICA ex3R
ATGGGGGGCACTGTTTCTCTCAGGACTA
26
HMICA ex4F
A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com