Method for extracting genome DNA of jellyfish
An extraction method, genome technology, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] The jellyfish polyp was picked up from the corrugated plate (or its attachment) with a toothpick, and collected into a 1.5ml centrifuge tube. Carefully shred with surgical scissors until there is no obvious granular tissue, add 400ul CTABbuffer, and set aside. Add 5ul proteinase K, digest at 55°C for 24 hours until the solution is clear and ready for use.
[0024] Then add the mixed solution of phenol, chloroform and isoamyl alcohol equal to the volume of the clarified solution, mix evenly and centrifuge at 12000rpm for 30min to suck the supernatant, then add the mixed solution of chloroform and isoamyl alcohol equal to the volume of the gained supernatant, and mix upside down Evenly centrifuge at 12000rpm for 30min to aspirate the supernatant. Add 0.7 times the volume of isopropanol to the obtained supernatant to precipitate at -20°C for 10 minutes, centrifuge at 12,000 rpm for 20 minutes, and discard the liquid. Wash twice with 500 ul of 70% ethanol at 12000 rpm for...
Embodiment 2
[0028] Collect the jellyfish sphenoids preserved in 95% alcohol into a 1.5ml centrifuge tube. Add 400ul homogenate buffer, 40ul 10% SDS, 2ul proteinase K, seal and digest at 55°C for 1 hour. After digestion, take out and add 400ul 6M NaCl, shake for 30 seconds and then centrifuge at 12000rpm for 30 minutes. Aspirate 800ul of the supernatant and add 560ul of isopropanol, precipitate at -20°C for 10 minutes, then centrifuge at 12000rpm for 20min, and remove the supernatant. Wash twice by centrifugation with 70% ethanol at 12,000 rpm for 5 minutes, and dry at room temperature. That is, the DNA of the jellyfish disc is obtained; the DNA of the jellyfish disc is stored in sterilized pure water at -20°C.
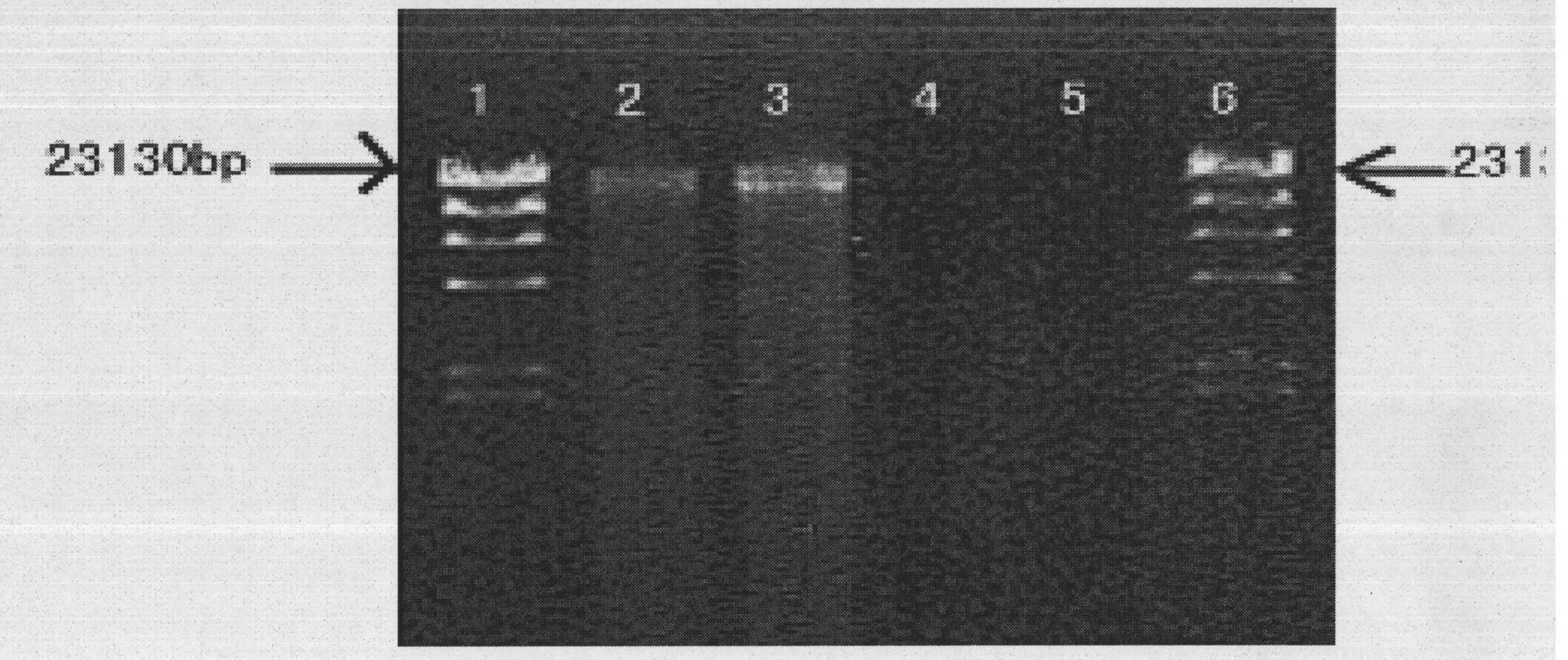
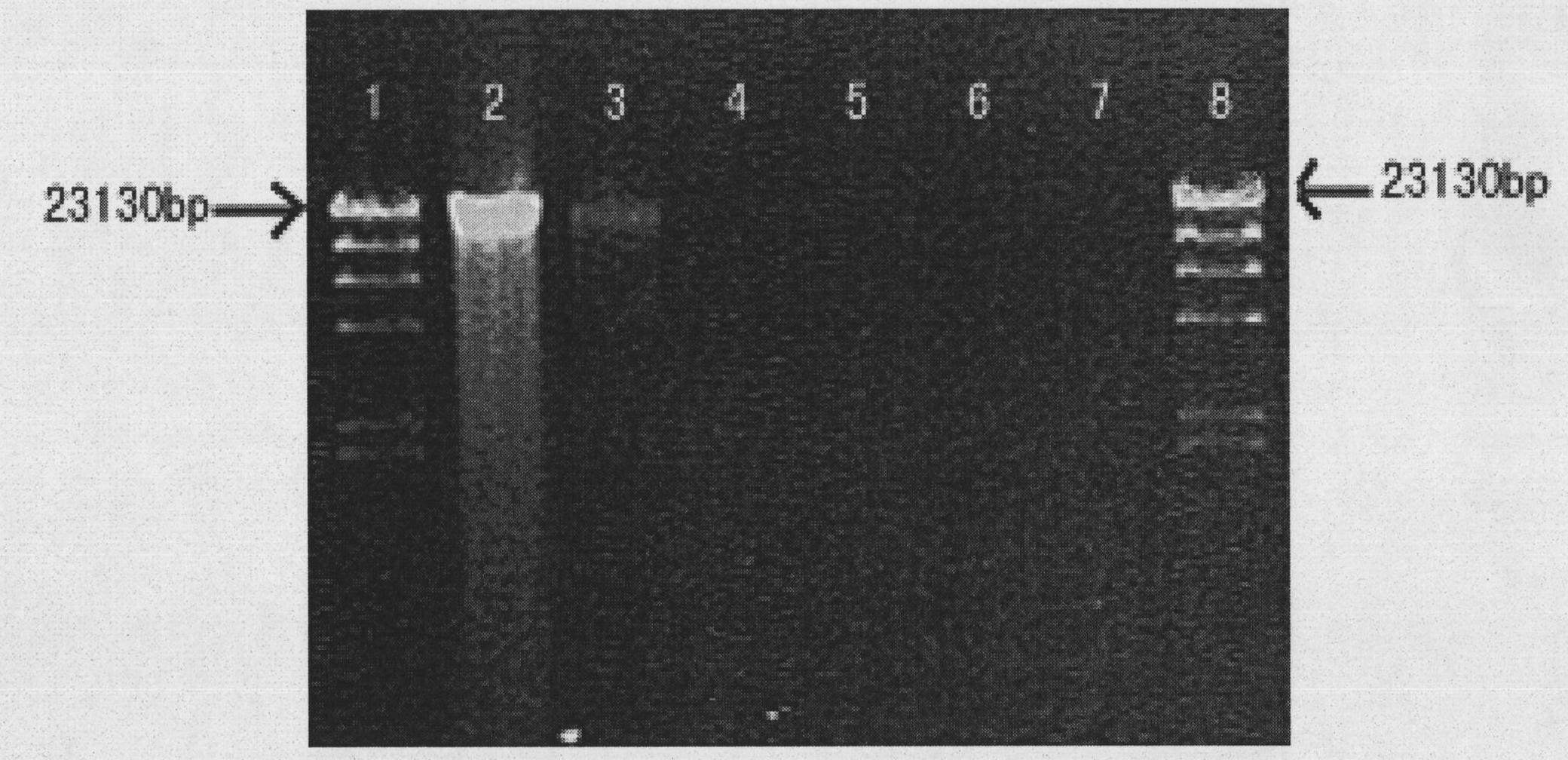
[0029] The DNA that obtains the jellyfish polyp is dissolved in 40ul sterilized pure water, and can be used for agarose gel electrophoresis detection after 30min (see figure 2 ).
[0030] The buffer solution contains 0.01M Tris-Hcl and 0.1M EDTA; the isopropanol and ethanol w...
Embodiment 3
[0032] The difference from Example 1 is:
[0033] Shred the jellyfish polyp with surgical scissors until there is no obvious granular tissue, add CTAB buffer 300ul, and set aside;
[0034] Then add 3ul of proteinase K with a concentration of 20mg / ml, digest at 56°C for 26 hours until the solution is clear, and set aside;
[0035] Add 1 times the volume of isopropanol to the supernatant after centrifugation with chloroform and isoamyl alcohol, precipitate at -18°C for 15 minutes, take out after precipitation, centrifuge, discard the liquid, and use a volume concentration of 70 % ethanol, centrifuged and washed once, and the excess liquid was poured out at room temperature and dried naturally to obtain the genomic DNA of the jellyfish polyp.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com