Method for culturing transgenic plant with decreased wax
A plant, coding gene technology, applied in the fields of botanical equipment and methods, biochemical equipment and methods, plant peptides, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1, discovery of gene
[0033] 1. Phenotype analysis of rice waxy mutant osgl1
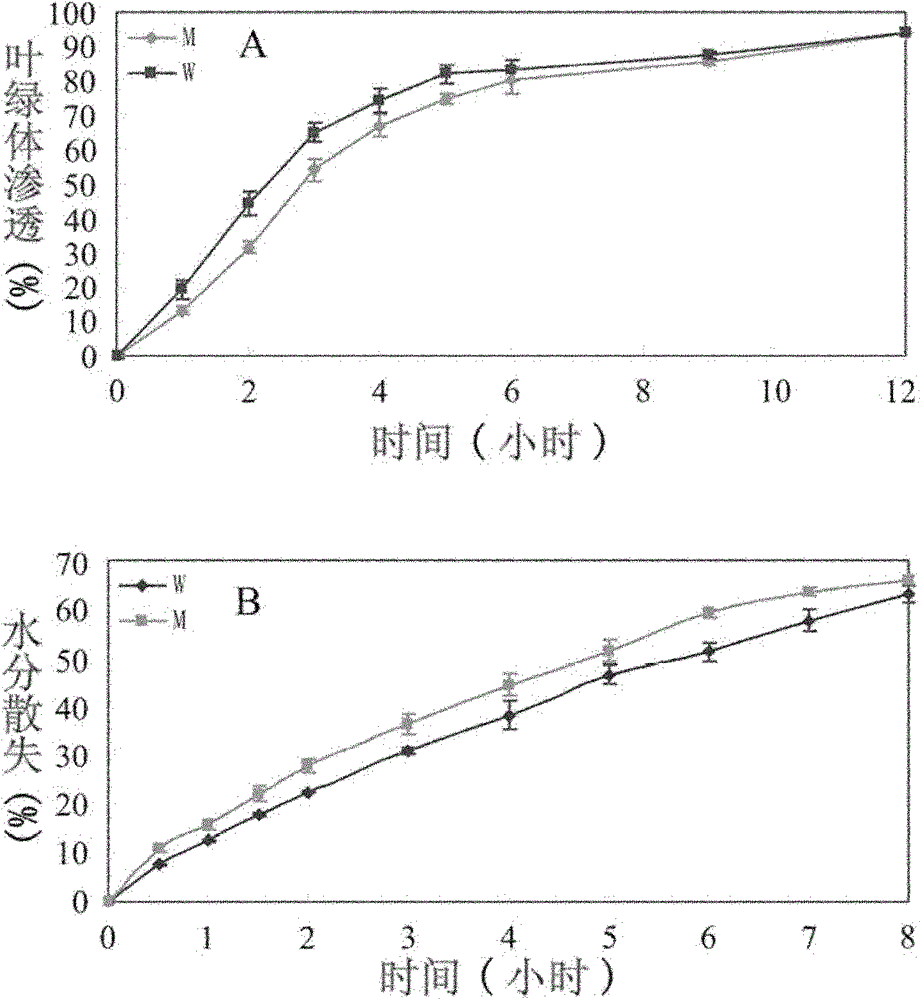
[0034] The rice waxy deletion mutant osgl1 is a natural mutant of wild-type 3037. Under normal conditions, mutant osgl1 had no significant difference compared with wild-type 3037 ( figure 1 A), but in the case of rain or artificial shower, the leaves of mutant osgl1 showed a highly hydrophilic phenotype. In wild-type 3037, due to the presence of a large amount of wax in the leaf epidermis, water molecules tend to aggregate on the leaf surface to form spherical water droplets. As long as the leaves are shaken a little, these water droplets on the leaves will drip. In the mutant osgl1, due to the high hydrophilicity of the leaves, water molecules will be evenly distributed on the entire leaf surface without forming water droplets ( figure 1 B). figure 1 Middle, wild type on the left, mutant on the right.
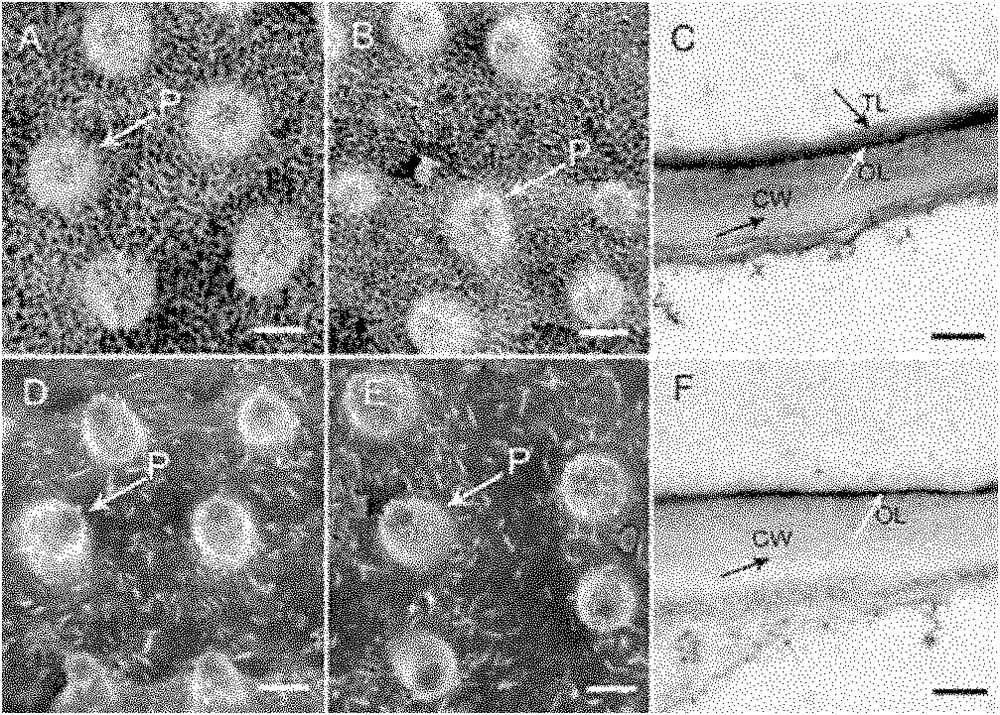
[0035] 2. Electron microscope observation and analysis of mutant osgl1 ...
Embodiment 2
[0045] Example 2, the acquisition of OsGL1 and its coding gene OsGL1
[0046] 1. Map-based cloning of the genome gene of OsGL1
[0047] In order to clone the OsGL1 gene, the homozygous mutant leaf waxy deletion mutant was crossed with Zhonghua 11, and the obtained F 1 F 2 In the population, 180 F2 recessive individuals (F2 individuals with leaf wax loss phenotype) were initially mapped for the OsGL1 gene. Using STS molecular markers and using PCR method, it was found that the STS markers P1, P2, P3, P4, P5 and P6 on chromosome 9 had obvious linkage with the mutation site. Most of the exchanged plants between the mutation site and P3 were also exchanged between the mutation site and P1 and P2, and most of the exchanged plants between the mutation site and P4 were included in the mutation site In the exchange individual plants between P5 and P6. At the same time, the exchange individual plants between the mutation site and P3 were different from the exchange individual plant...
Embodiment 3
[0062] Embodiment 3, the application of gene OsGL1
[0063] 1. Complementation experiment of waxy deletion mutant osgl1 phenotype
[0064] 1. Construction of complementary vector pCGL and complementary control vector pCGLC
[0065] BAC OSJNBb0057D06 was digested with Xbal to obtain a full-length DNA fragment (8264bp) comprising 2899 bases upstream of the start codon ATG of OsGL1 and 1436 bases after the stop codon TGA, and cloned into pCAMBIA1300 (CAMBIA , Canberra, Australia) between the Xbal restriction sites to obtain the recombinant vector; digest the constructed complementary vector pCGL with HindIII, remove part of the coding region of OsGL1 and the 5' promoter region of the gene, and retain part of the coding region at the 3' end and the 3' regulatory region, that is, the complementary control vector pCGLC was constructed ( Figure 5 ).
[0066] pCGL, complementary expression vector, including 2899bp upstream and 1436bp downstream of OsGL1 gene;
[0067] pCGLC, the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com