Interferon response in clinical samples (IRIS)
A technology for interferon beta and biological samples, which is applied in the direction of material testing products, microbial determination/inspection, biochemical equipment and methods, etc., and can solve problems such as no cure.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
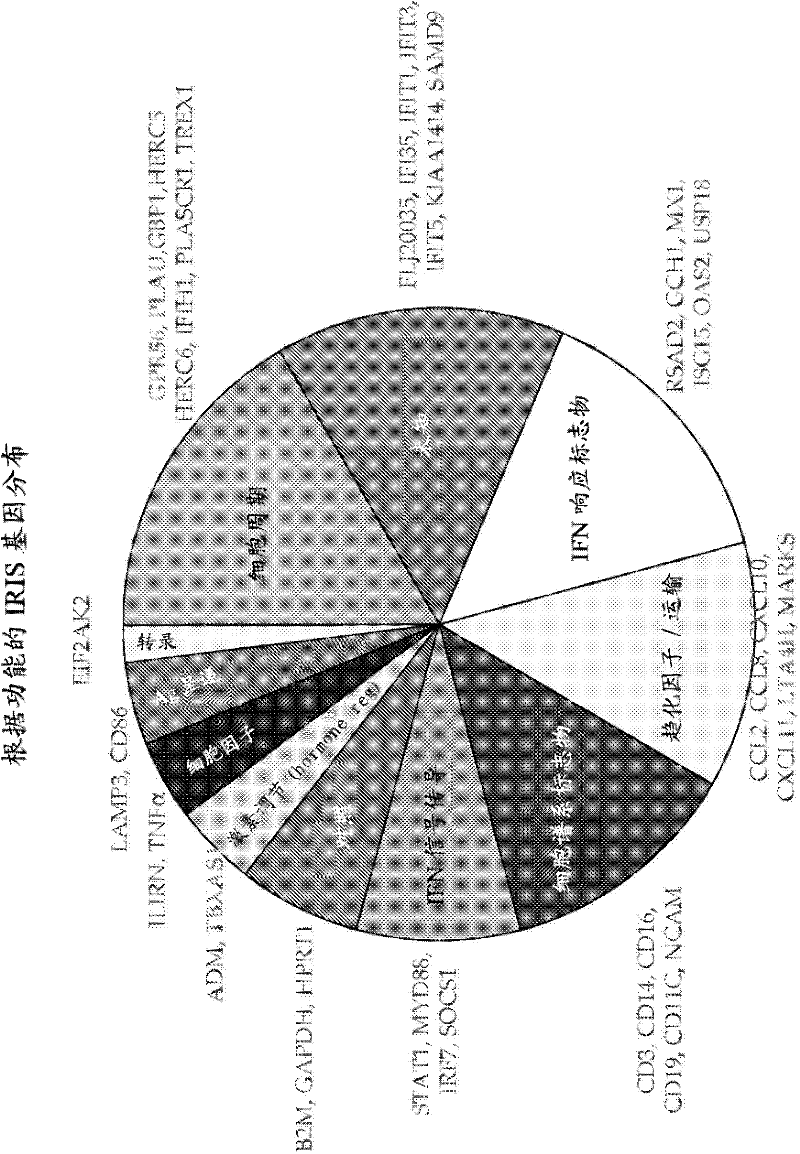
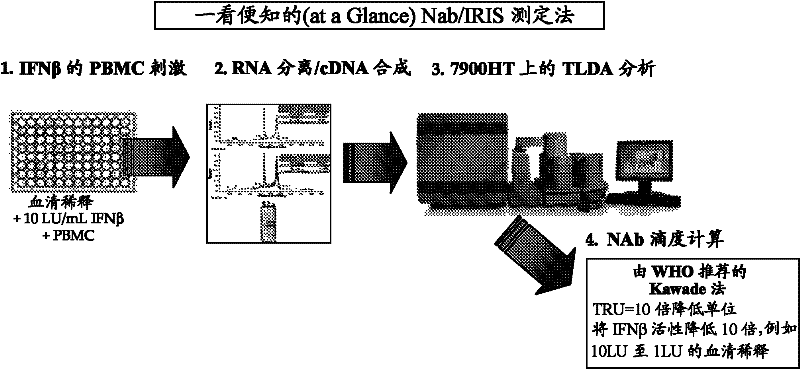
[0125] Example 1: IRIS gene expression analysis
[0126] Relative gene expression is measured with a sequence detection system (eg ABI Prism 7900HT) using application software (eg SDS2.1) designed for the detection unit. Using Equation 2- δδCTThe expression level of each IRIS gene was calculated by the comparative CT method, where δδCT equals the normalized signal level in sample "A" (eg, IFNβ-stimulated) relative to the normalized signal level in a calibration sample (eg, non-stimulated control). Samples can be normalized using the GAPDH or HPRT1 housekeeping genes. Alternatively, when examining responses in patient PBMC samples, T cells (CD3), B cells (CD19), monocytes (CD14), dendritic cells (ITGAX), neutrophils (NCAM), or NK cells can be used Samples were normalized for cell lineage markers of cells (CD16). For Nab analysis, IRIS gene expression was compared between samples containing patient serum concentrations with 10 LU / mL IFNβ and calibration samples with only 10 L...
Embodiment 2
[0127] Example 2: Nab Assay Patient Data
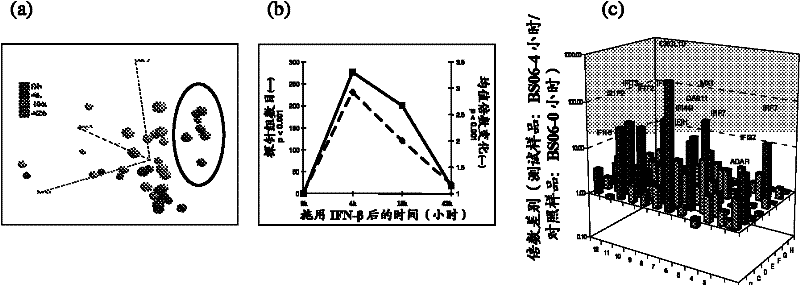
[0128] Using the IRIS gene expression analysis described above, we found that the degree of neutralization of IFNβ induction appears to be unique for each IRIS gene analyzed. For example, the standard IFNβ-responsive gene MxA gene is very sensitive to neutralization, whereas other IRIS genes require higher serum neutralization concentrations. (see Figure 6 ). This was shown in an analysis of patient sera previously characterized by viral inhibition assays as having potent Nab activity. Furthermore, sensitivity to neutralization (as indicated by a gene's TRU titer) did not correlate with the magnitude of the response for that particular gene. (see Figure 7 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com