Construction method of novel biological network model
A biological network and construction method technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve problems such as difficult to reflect the characteristics of protein domain reorganization networks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
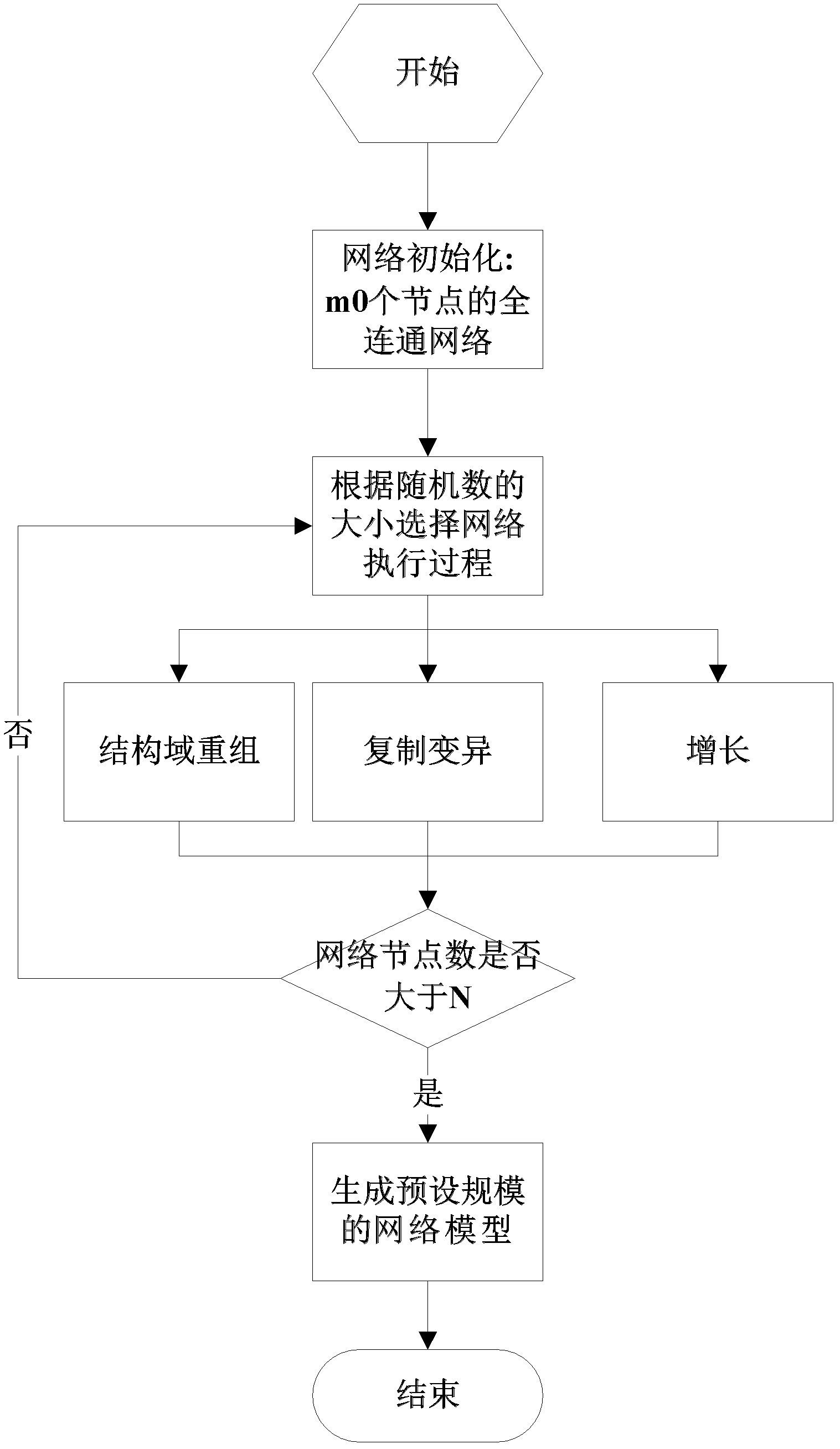
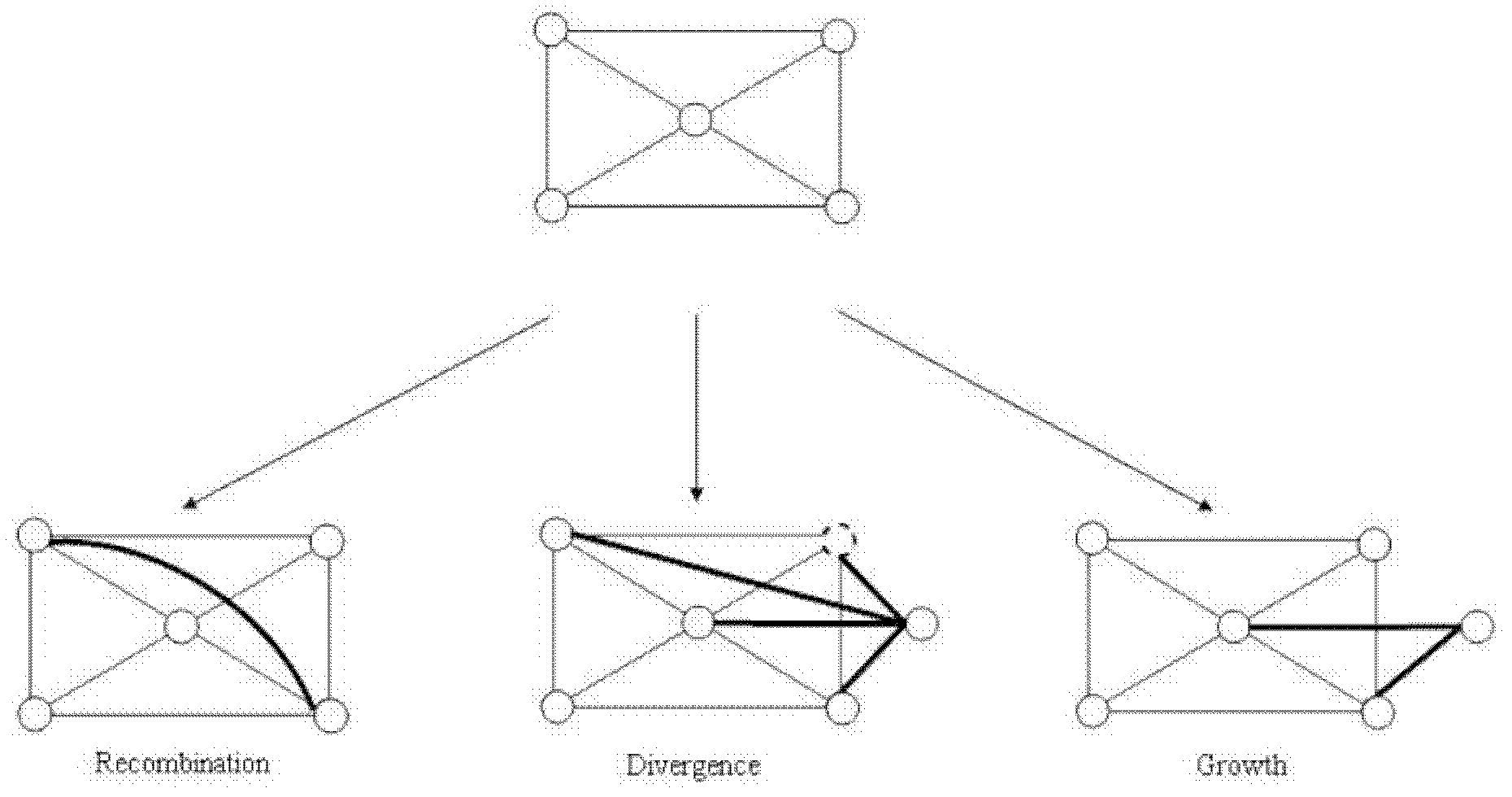
[0051] Such as figure 1 As shown in ~3, the biological network model is a network constructed by simulation in a computer environment. figure 1 It is a flow chart of the steps of the complex network evolution model modeling method of the present invention. Such as figure 1 As shown, firstly, a small-scale fully connected network with the number of nodes m0 is generated (for example, a fully connected network with m0=3, where nodes are interconnected in pairs by edges). Then in unit time, the network evolves according to the following rules: the newly added node chooses one of the reorganization, mutation or growth process with different probability r. For specific network changes, see figure 2 , That is, when 0<r<R, select the recombination process, when R<r<(R+D), choose the mutation process, when (R+D)<r<1, choose the growth process; when the size of the network reaches the preset When the scale of the network is over, the network modeling process ends.
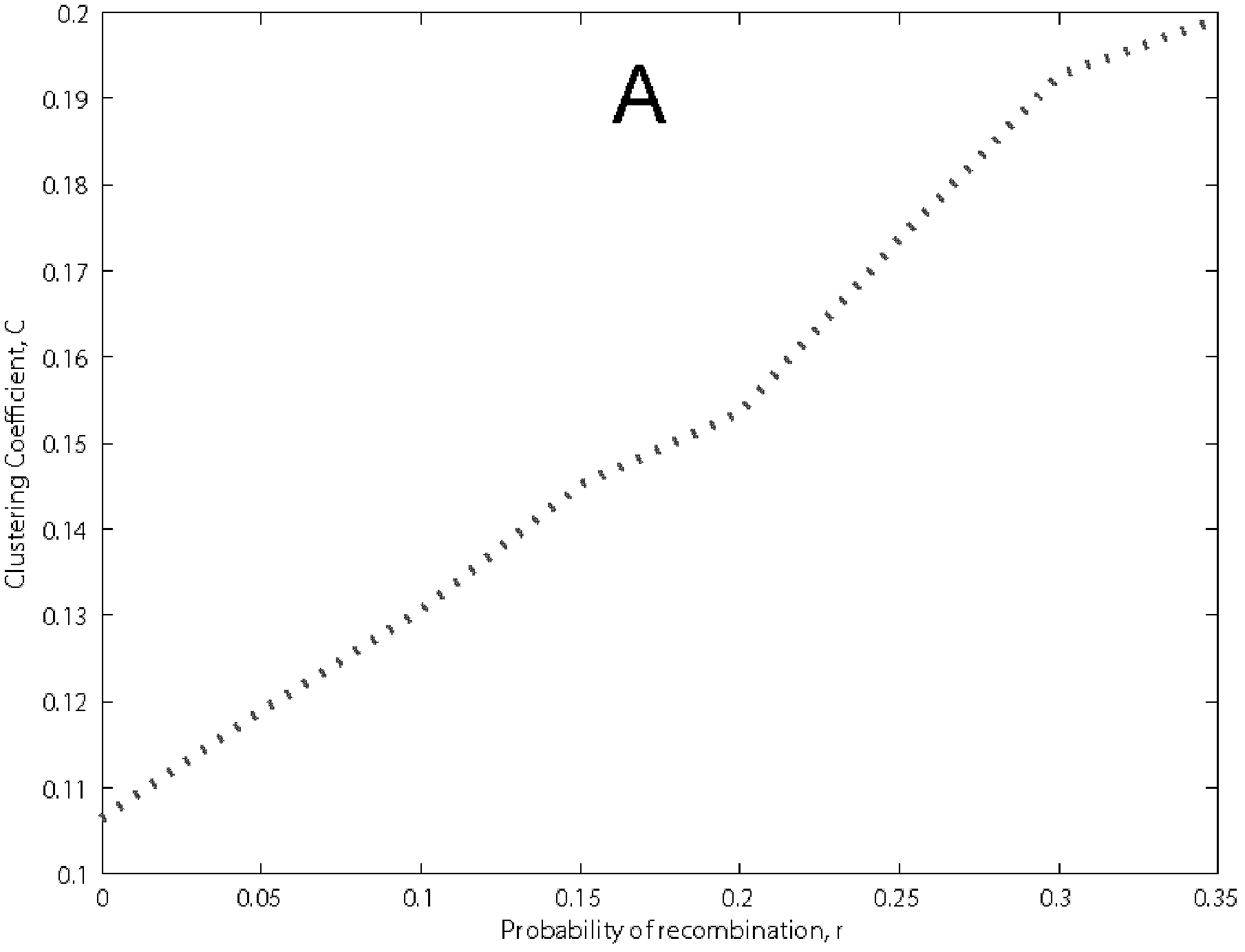
[0052] Figure 3 show...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com