Massively parallel continguity mapping
A technology of adjacency information and adapters, applied in the field of D, can solve problems such as the astonishingly high price of DNA sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
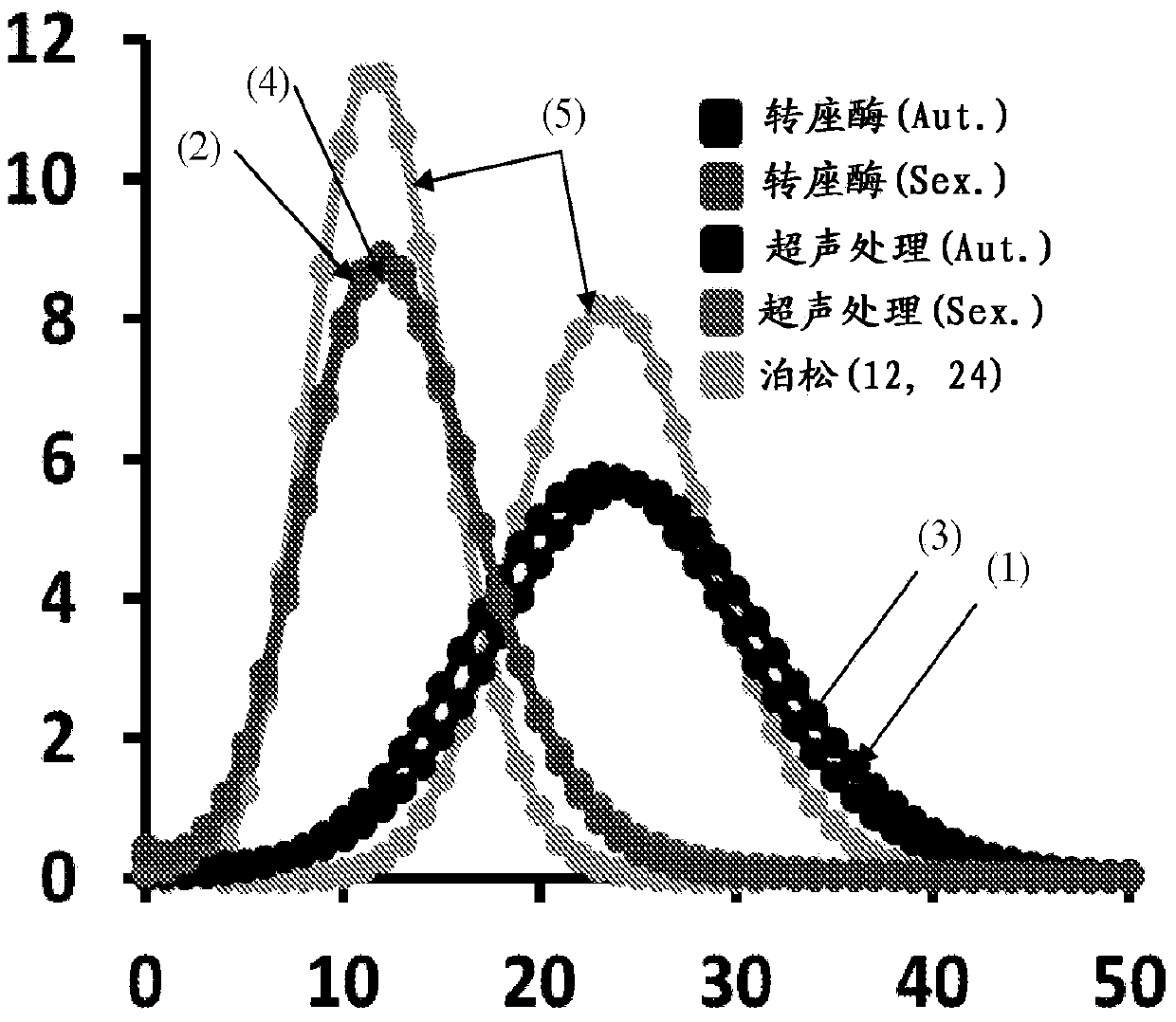
[0099]Several properties of in vitro transposition can be exploited to develop ultra-low-cost, massively parallel sequencing methods for capturing contiguity information at different scales. First, the modified Tn5 transposome attacked DNA in vitro with high efficiency and density in a reaction that catalyzed the insertion of consensus sequences, with or without breaks, depending on whether the synthetic transposon was continuous or discontinuous. Second, the pattern of transposome attack is relatively random with respect to sequence content. Third, degenerate subsequences plus consensus adapter sequences can be easily included in synthetic transposons. Fourth, in vitro transposition is inexpensive as a one-volume, aqueous-phase, enzymatic reaction. Examples 1-3 relate to the development of massively parallel methods using in vitro transposition to inform small, medium and large range contiguities, respectively. Example 4 concerns the development of a method for capturing co...
example 6
[0099]Several properties of in vitro transposition can be exploited to develop ultra-low-cost, massively parallel sequencing methods for capturing contiguity information at different scales. First, the modified Tn5 transposome attacked DNA in vitro with high efficiency and density in a reaction that catalyzed the insertion of consensus sequences, with or without breaks, depending on whether the synthetic transposon was continuous or discontinuous. Second, the pattern of transposome attack is relatively random with respect to sequence content. Third, degenerate subsequences plus consensus adapter sequences can be easily included in synthetic transposons. Fourth, in vitro transposition is inexpensive as a one-volume, aqueous-phase, enzymatic reaction. Examples 1-3 relate to the development of massively parallel methods using in vitro transposition to inform small, medium and large range contiguities, respectively. Example 4 concerns the development of a method for capturing co...
example 1
[0107] Example 1: Small-scale adjacency
[0108] 1.A. Symmetrically and Uniquely Mark Fragmentation Events
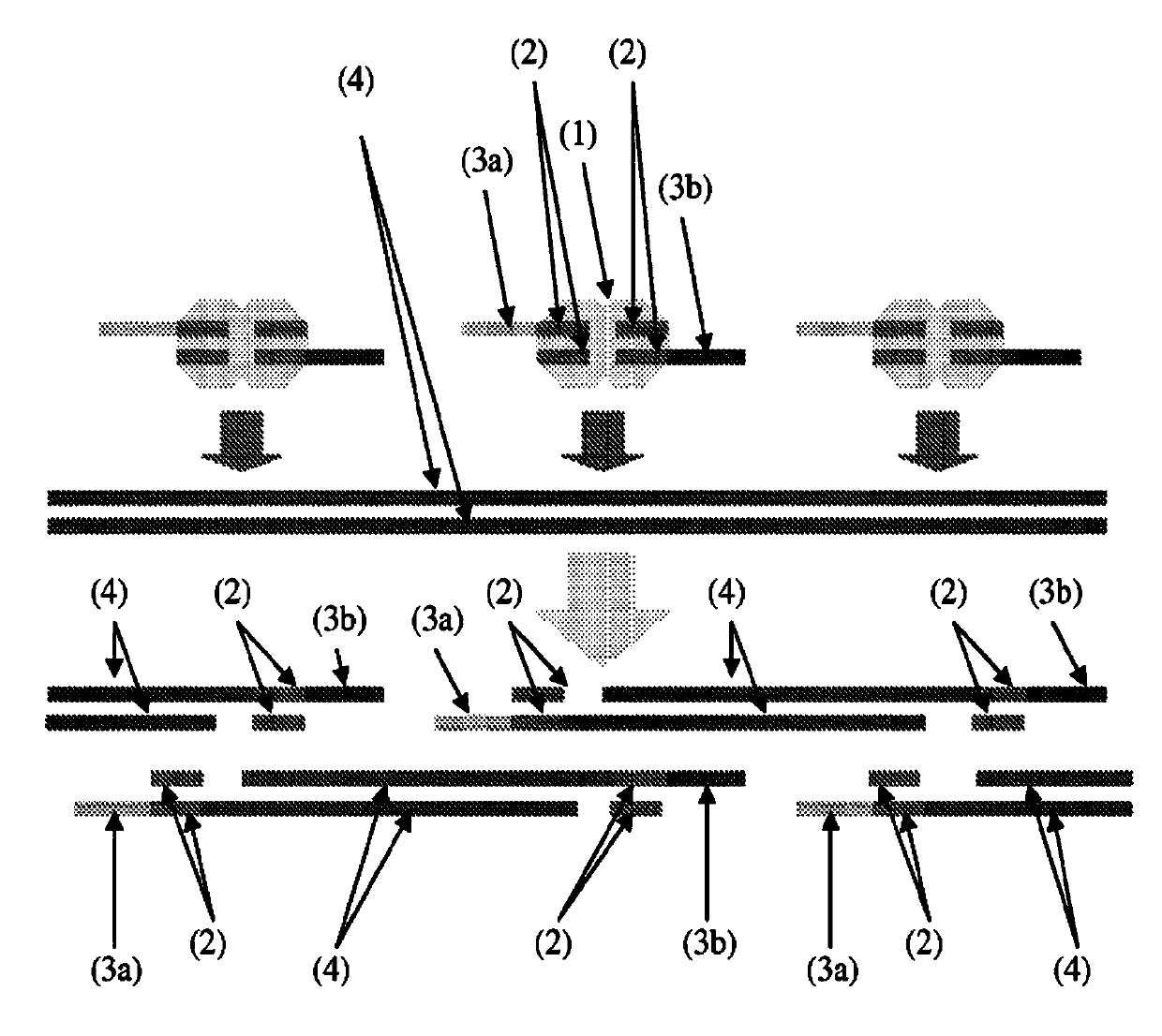
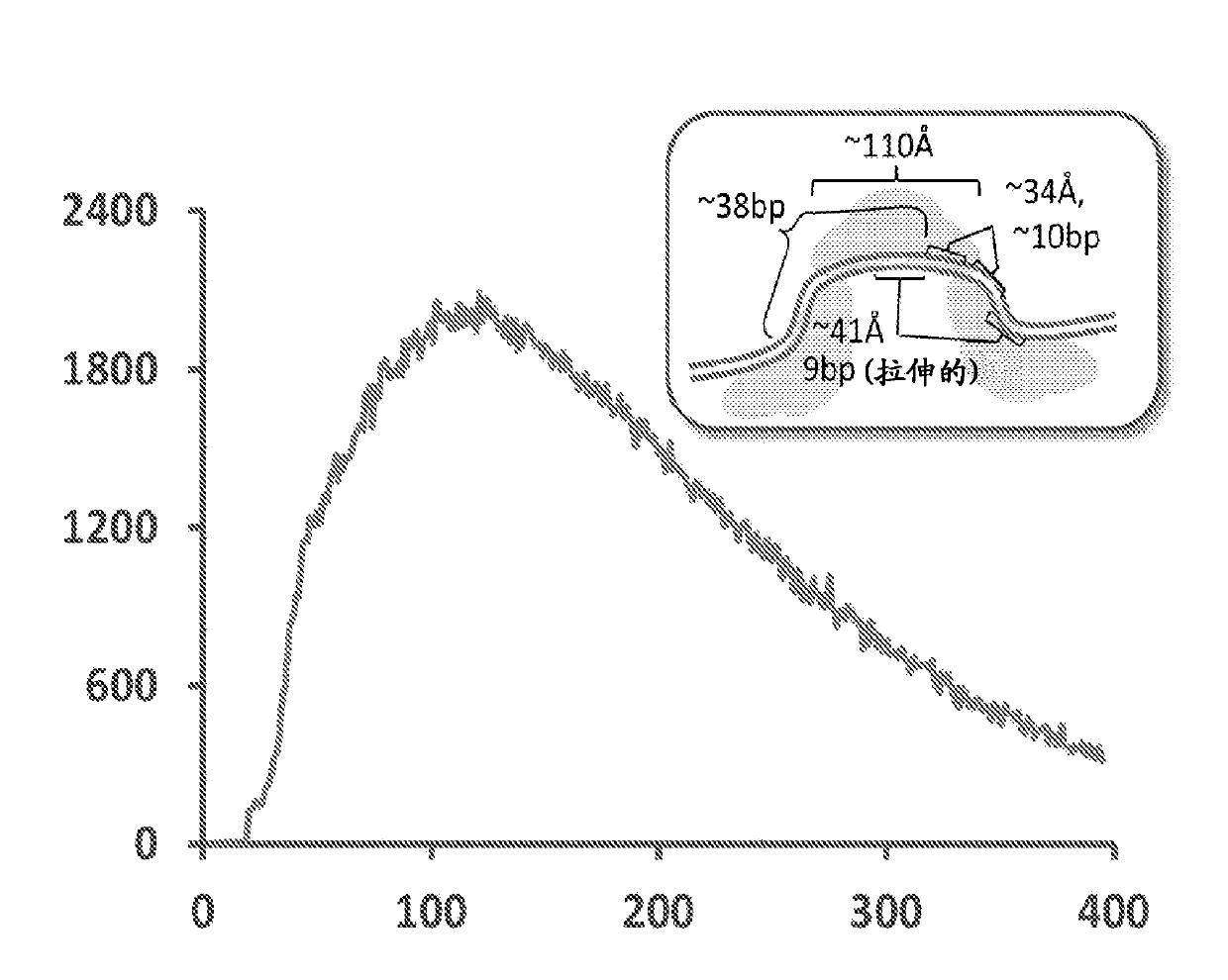
[0109] Genomic DNA breaks, whether by mechanical or enzymatic means, result in a complete loss of information about the pairing of molecules originating from either side of any individual "break". To preserve this information, methods were designed so that unique barcodes were bound to both ends of fragments derived from each disruption introduced by in vitro transposition ( Figure 4 ). Briefly, transposases can be used to catalyze the in vitro insertion of synthetic transposons containing nicking restriction endonucleases into very low amounts of genomic DNA, i.e., less than 5 haploid human genome equivalents. A degenerate single-stranded "bubble" flanking the site. and figure 1 In contrast to the method described in , the synthetic transposon is contiguous, containing a 19 bp ME sequence together with two endonuclease nicking sites flanked by 25 bp of degenera...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com