Astaxanthin synthase of Sphingomonas and its encoding gene and method for genetic manipulation of Sphingomonas
A sphingomonas, coding gene technology, applied in the field of genetic manipulation, astaxanthin biosynthetic enzyme and its coding gene, can solve the problem of no genetic manipulation, etc., to achieve the effect of enriching gene diversity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
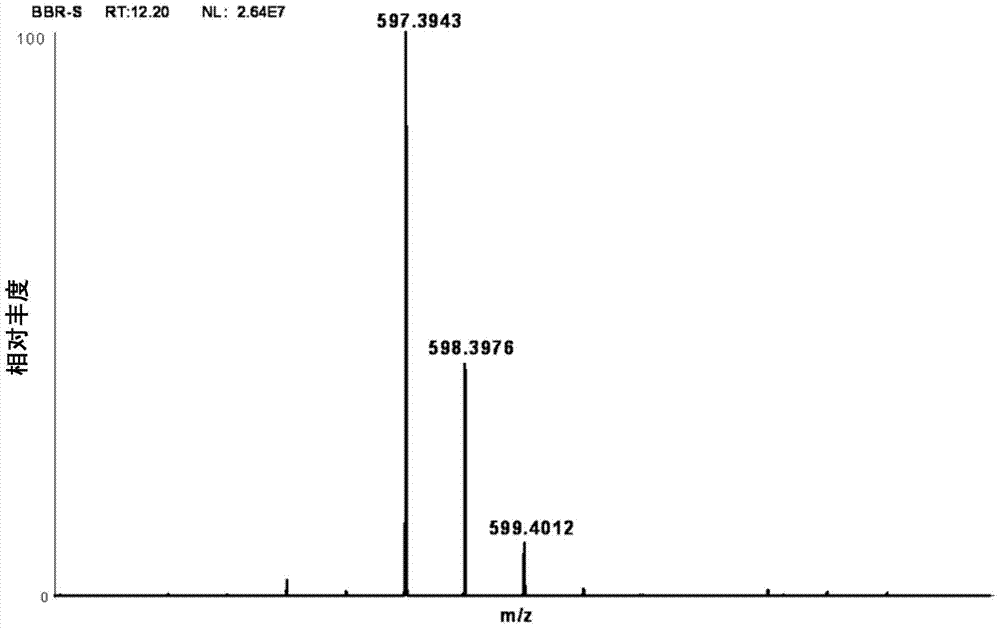
[0044] Example 1 Detection of Astaxanthin Products in Sphingomonas
[0045] Extract Sphingomonas Sphingomonas ATCC55669 (purchase from ATCC) metabolites, the method is as follows:
[0046] The strains were activated on a #272 medium plate (nutrient broth 8g / L, glucose 5g / L, agar 1.6%) and cultured at 26°C. Pick the colony and culture it in 5mL #272 medium (nutrient broth 8g / L, glucose 5g / L) at 26°C and 220rpm. After 24 hours, transfer to 100mL#272 medium for culture at 26°C and 220rpm, OD 600 At 0.8, transfer to 300mL #272 medium for culture at 26°C and 220rpm, and harvest the bacteria after 60h.
[0047] Centrifuge the bacterial solution at 8000rpm for 10min, collect the bacterial cells, add 10mL extractant (V 丙酮 :V 甲醇 =4:1), shake to break up the cells, extract for 10min, centrifuge at 8000rpm, 4°C for 5min, remove the supernatant, and extract 3 times according to the above steps, collect the supernatant, spin dry, add 3mL acetone to dissolve, and centrifuge at 13000rpm f...
Embodiment 2
[0048] Example 2 Determination of related astaxanthin biosynthesis genes in Sphingomonas
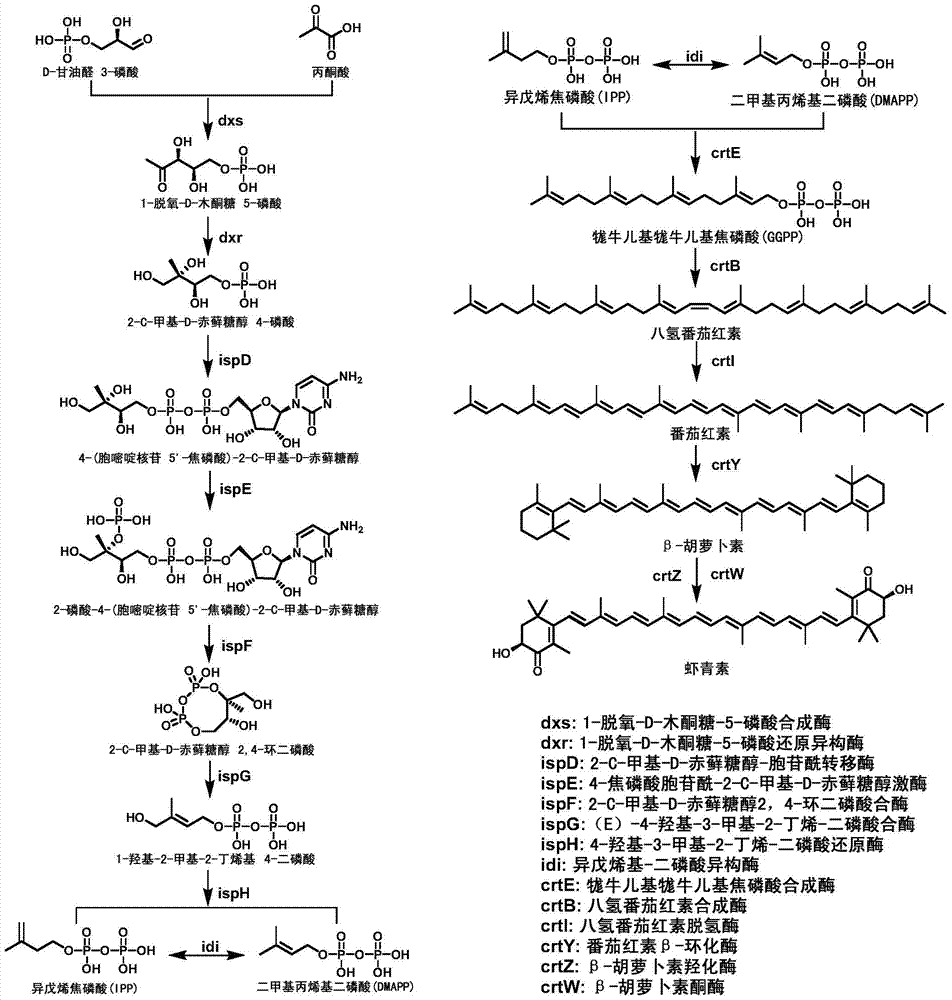
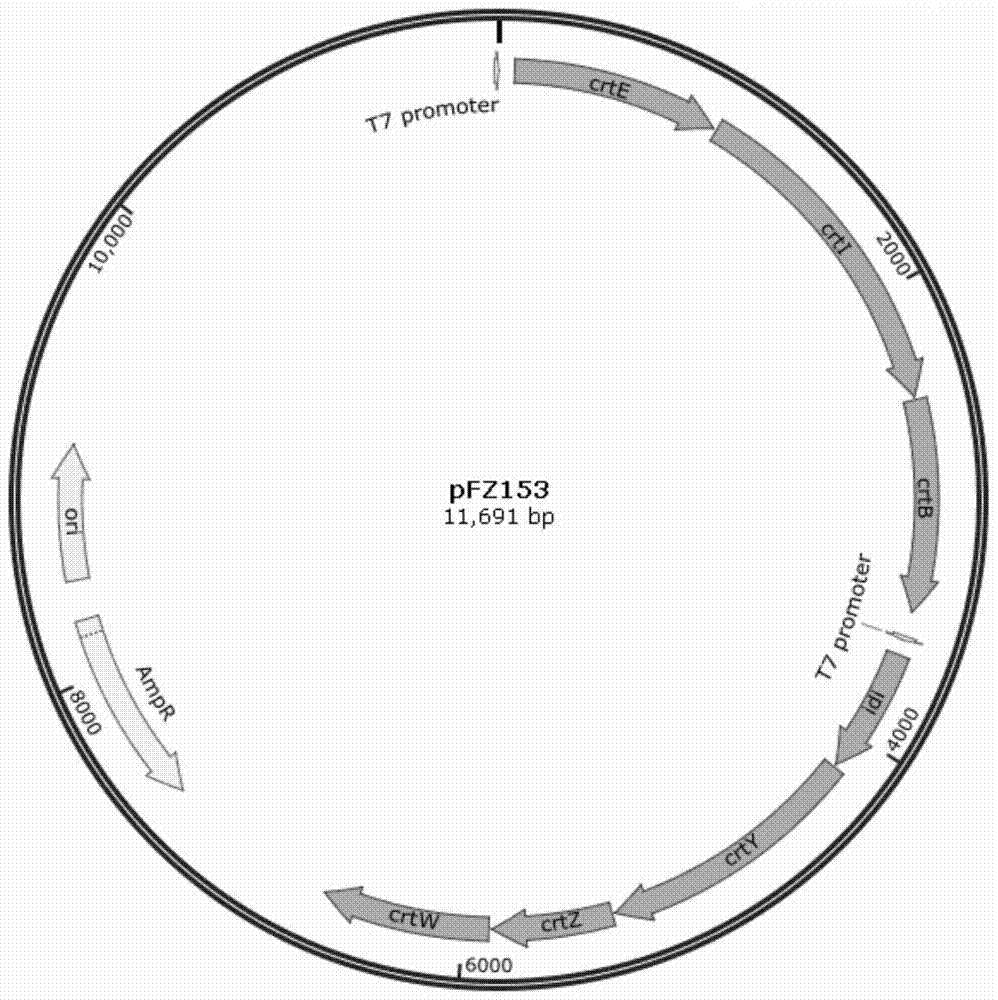
[0049] As can be seen from Example 1, Sphingomonas Sphingomonas ATCC55669 can produce astaxanthin, and this strain contains a biosynthetic pathway capable of producing astaxanthin. Comparing the genome information of Sphingomonas in NCBI (http: / / www.ncbi.nlm.nih.gov / ) Blastp, it was found that there is a MEP pathway in Sphingomonas, which starts from pyruvate IPP and DMAPP were synthesized via dxs, dxr, ispE, ispDF, ispG, ispH. IPP and DMAPP generate astaxanthin through the carotenoid synthesis pathway through crtE, crtB, crtI, crtY, crtZ, and crtW. In this linear pathway of astaxanthin synthesis by Sphingomonas, all the other genes except crtE and crtZ are unique.
Embodiment 3
[0050] Example 3 Amplification of Sphingomonas crtE gene and crtZ gene
[0051] Genomic DNA was extracted from Sphingomonas Sphingomonas ATCC55669, and the extraction method was as follows:
[0052] (1) Take 50mL of fresh bacterial solution to a conical centrifuge tube, centrifuge at 7000rpm×5min, and discard the supernatant.
[0053] (2) Add 10mL of ddH 2 O, break up on the shaker, centrifuge at 7000rpm×5min, and discard the supernatant.
[0054] (3) Add 10 mL of SET buffer (75 mM NaCl, 25 mM EDTA, 20 mM Tris-Cl), disperse on a shaker, centrifuge at 7000 rpm for 5 min, and discard the supernatant.
[0055] (4) Add 5 mL of SET buffer, disperse it on a shaker, add 150 μL of lysozyme (lysozyme, 100 mg / mL, -20°C), bathe in 37°C water for 30-60 minutes, shake slowly every 5-10 minutes, Until the cell wall is completely lysed (to identify the cell wall is completely lysed: take a small amount of bacterial liquid, add 1 drop of 10% SDS, the bacterial liquid is clear and silky). ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com