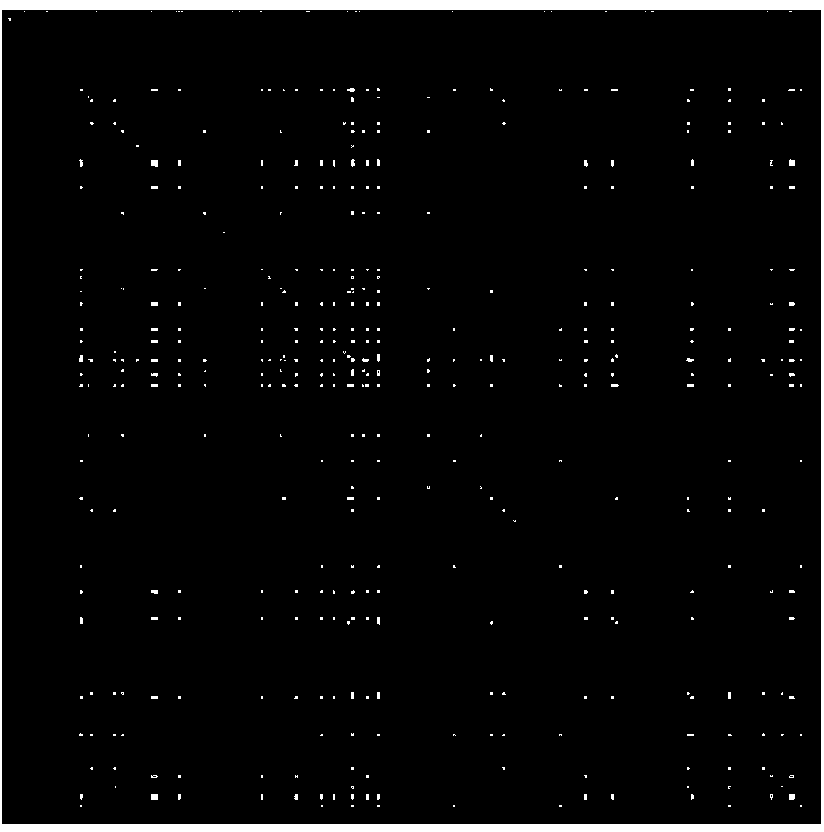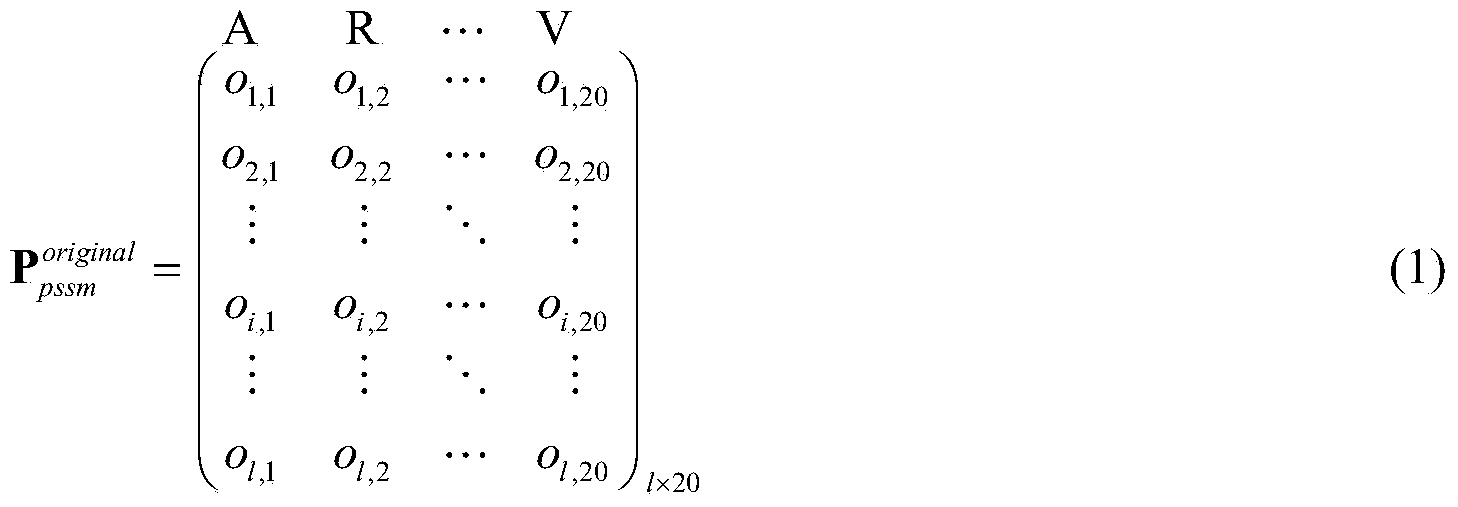GPCR(G Protein-Coupled Receptor)-drug interaction prediction method based on postprocessing study
A technology of coupling receptors and interaction, applied in special data processing applications, electrical digital data processing, instruments, etc., can solve the problems of large gap between prediction accuracy and actual application, lack of related information, poor interpretability, etc. Prediction speed and accuracy, and the effect of improving interpretability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0052] In order to better understand the technical content of the present invention, specific embodiments are given together with the attached drawings for description as follows.
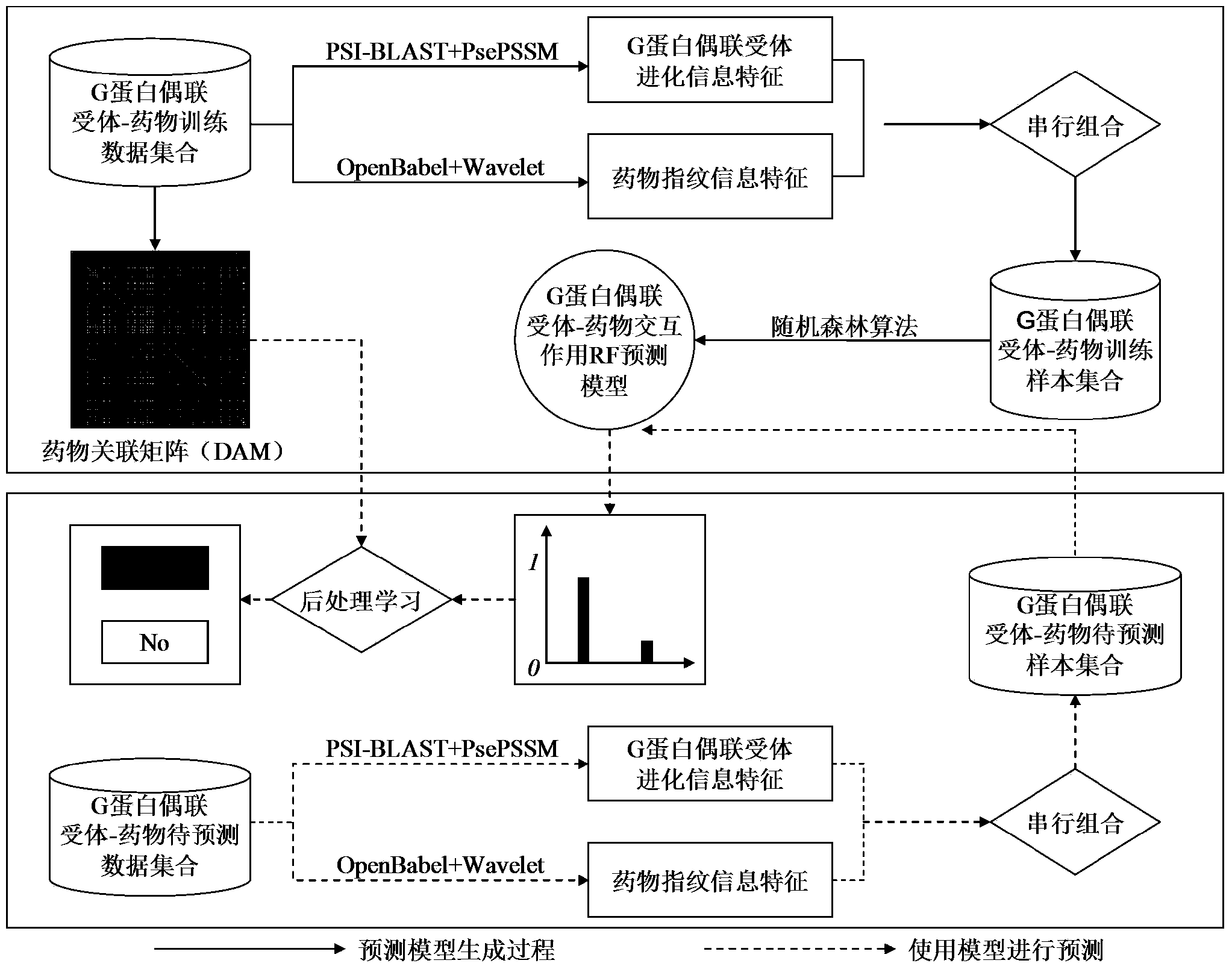
[0053] Such as figure 1 As shown, according to a preferred embodiment of the present invention, a G protein-coupled receptor-drug interaction prediction method based on post-processing learning, its realization includes the following steps:
[0054] Step 1: Based on the information of all G protein-coupled receptors with interactions (i.e., positive samples) in the training data set, construct a formula that describes the probability that different drugs can bind to the same G protein-coupled receptor Drug Association Matrix (DAM);
[0055] Step 2: Based on the input G protein-coupled receptor sequence information and drug molecular structure information, perform multi-view feature extraction and feature combination, that is, use the PSI-BLAST and PsePSSM algorithms to extract the evolution inform...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com