A method and its application of one-time accurate quantification of alkane hydroxylase gene alkb
An alkane hydroxylase, sub-accurate technology, applied in the field of molecular ecology and biology, can solve the problems of poor accuracy and comparability of quantitative results, differences in primer amplification efficiency, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
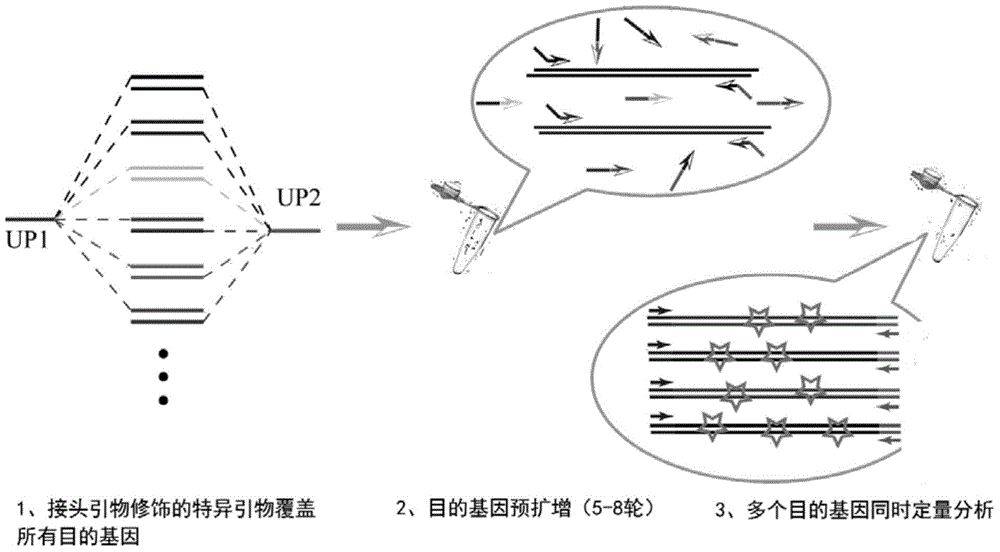
[0040] Example 1: Quantitative method for real-time fluorescent PCR with multiple primers based on universal sequence adapters
[0041] Step 1: Design and specificity verification of universal primers
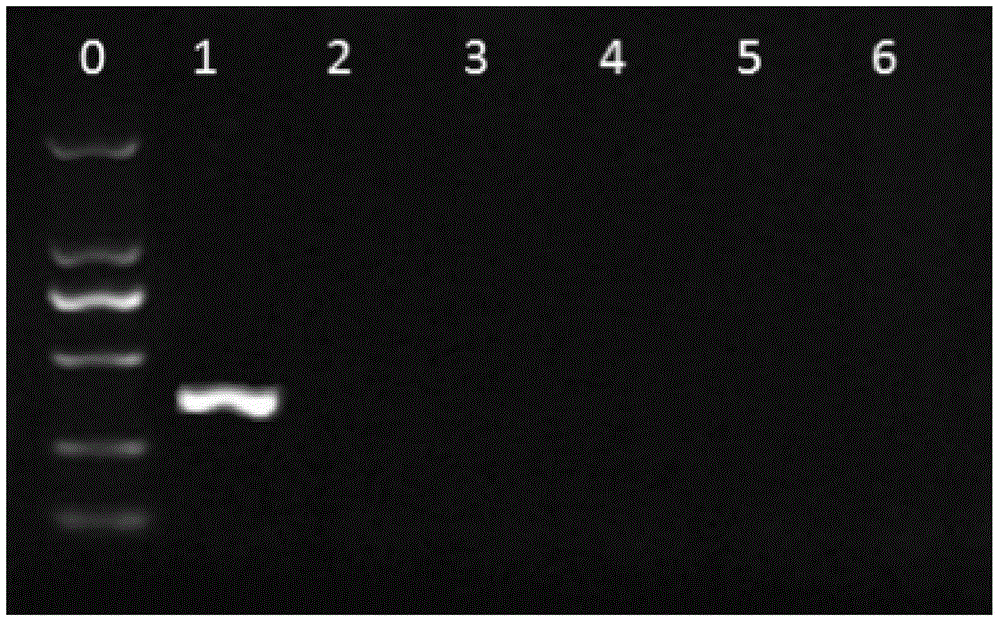
[0042] Randomly combine stop codons and rare codons of Escherichia coli into a primer sequence of about 20bp, use 6.0, Primer Premier 5.0 and other primer analysis software evaluate the quality of primers, and carry out comparison analysis in Genbank database, therefrom preferred two primer sequences UP-1 (SEQ ID No.1), UP-2 (SEQ ID No. 2) As a general sequence for multiplex real-time fluorescent quantitative PCR. Use this universal sequence to test a microbial genome positive quality control ( figure 2 , 1 lane) and 5 randomly selected environmental gene samples ( figure 2 , lanes 2 to 6) for ordinary PCR amplification, the amplification results showed that the universal primers had no non-specific amplification on environmental genome samples, and basically no primer-di...
Embodiment 2
[0078] Example 2: Comparison of multiple primer PCR amplification effects based on universal sequences and common multiple primer PCR
[0079] When performing multiple PCR amplification, the PCR process is performed with reference to the multiple real-time fluorescent quantitative PCR method based on the universal sequence provided in Example 1 of the present invention, except that the primers used in common multiple PCR are specific primers that remove the universal sequence linker, and No universal sequences were added as primers during PCR. The amplification results of templates containing the same amount of 7 alkB genes showed that the amplification efficiencies of different alkB genes by ordinary multiplex PCR were quite different, and even some alkB genes were not amplified effectively under the mutual inhibition of primers ( Figure 5 , lanes 1 to 7); and the multiplex PCR method based on universal primers provided by the present invention has substantially the same amp...
Embodiment 3
[0080]Example 3: Comparison of the coverage of clone libraries prepared by multiple primers based on universal sequence adapters and degenerate primers
[0081] The designed universal sequence adapter specific primers UJSPs and degenerate primer alkBwf / r were used to amplify the alkB fragment from hydrocarbon oxidizing bacteria and non-hydrocarbon oxidizing bacteria respectively, and the amplification effects of the two methods were compared.
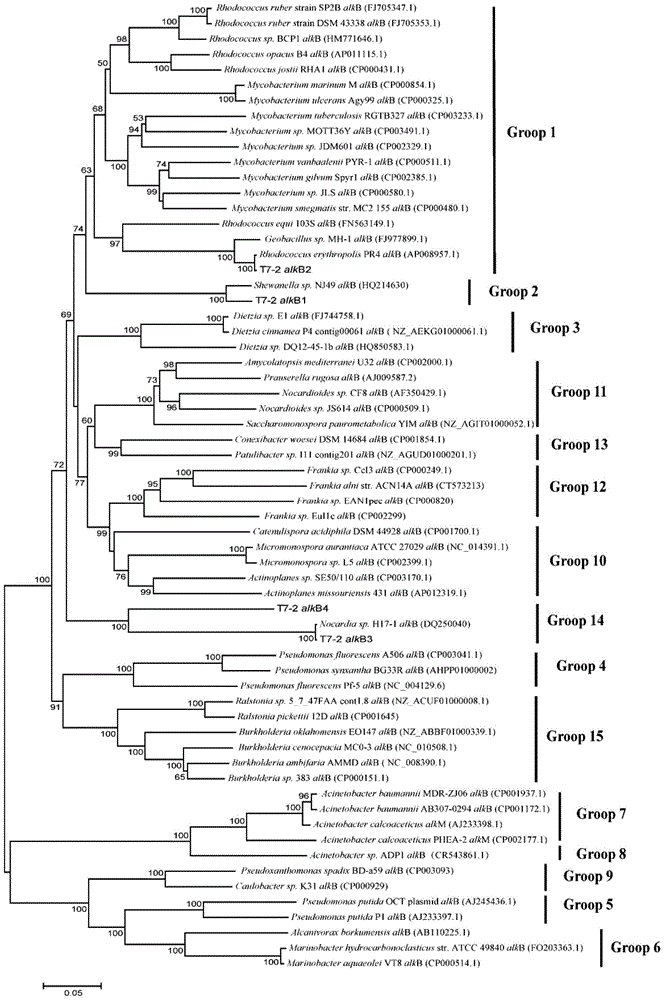
[0082] Formation water samples from oil reservoirs and 9 strains of hydrocarbon oxidizing bacteria (Marineella MCCC 1A00091, Acinetobacter haemolyticus CCTCC NK2.BH-7, Acinetobacter haemolyticus CCTCC NK2. Rhodococcus CGMCC4.1818, Rhodococcus roseus CGMCC 4.1480, Rhodococcus erythropolis CCTCC NK2.53968, Rhodococcus erythropolis CCTCC NK2.53968, Rhodococcus erythropolis NK2.CCTCC T7-2, Rhodococcus erythroflates CCTCC NK2.T1- 3) alkB fragment was amplified in a mixed bacterial sample composed of two strains of non-hydrocarbon oxidizing b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com