Application and detection method of single-stranded nucleotide sequence of inha gene as animal superovulation molecular marker
A single-stranded nucleotide and superovulation technology, which is applied to the application and detection field of animal superovulation molecular markers, can solve the problems of large sample consumption, large workload and high cost, and achieves convenient implementation, improved efficiency, and improved methods. easy effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] 1. Genomic DNA was extracted from the blood of Xiangxi yellow cattle with the Genomic DNA Extraction Kit of Tiangen Biochemical Technology Co., Ltd.
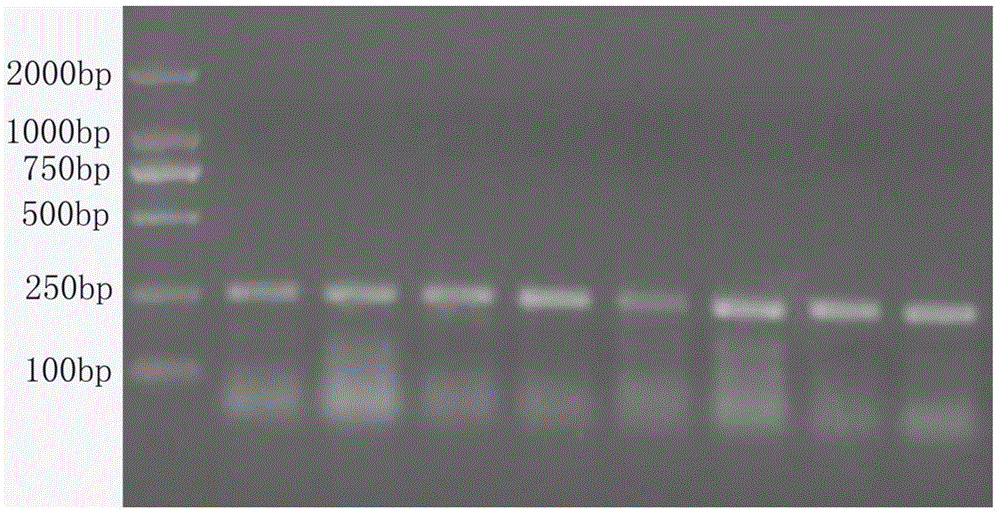
[0051] 2. Partial fragment amplification of INHA gene of Xiangxi yellow cattle:
[0052] Primers:
[0053]
[0054] PCR process:
[0055]
[0056]
[0057] For PCR results, see figure 1 .
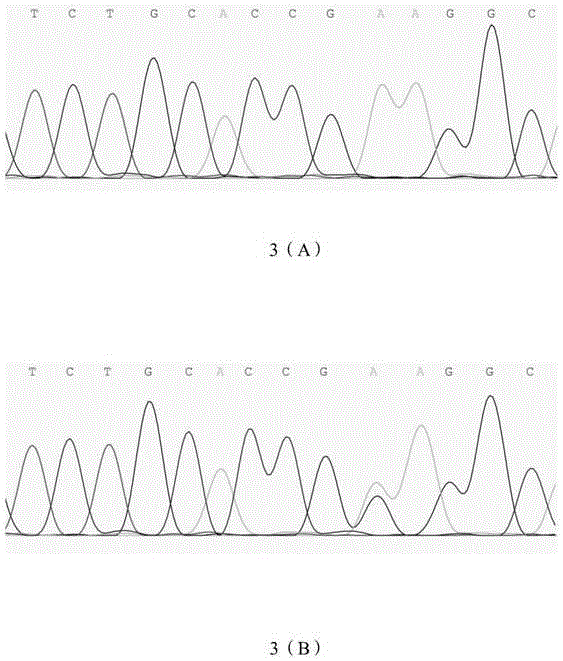
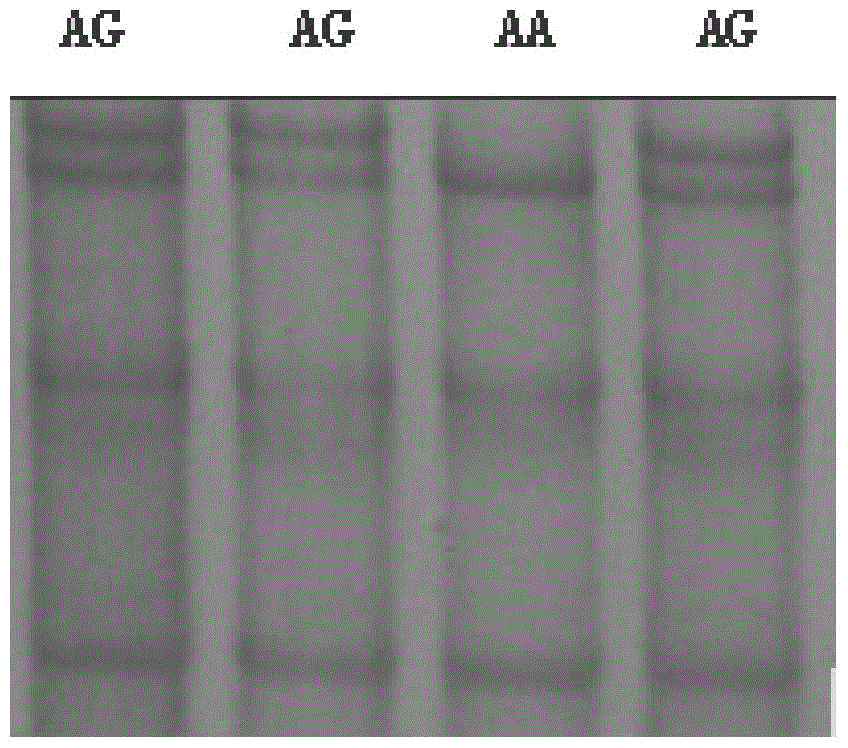
[0058] 3. SSCP typing of PCR amplification products of different individuals:
[0059] Mix 3 μl of PCR product with 7 μl of denaturing buffer (98% deionized formamide, 10 mmol / L EDTA (pH8.0), 0.025% xylene cyanol FF, 0.025% bromophenol blue), and use a PCR instrument 95 Denature at ℃ for 10 minutes, and then stand on ice for 5 minutes to avoid renaturation of the denatured product, thereby obtaining denatured single-stranded DNA. The denatured DNA was electrophoresed in a 12% non-denaturing polyacrylamide gel at 140V in a refrigerator at 4°C for 14h. The gel was silver-stained with 0.4% silver nitrate to develop the colo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com