Parallel accelerating method for BWT index construction for multiple sequences
A multi-sequence and indexing technology, applied in the field of bioinformatics, can solve the problems of slow and inefficient BWT index construction, achieve the effects of reducing the required time, improving the construction process, and being easy to transplant and promote
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
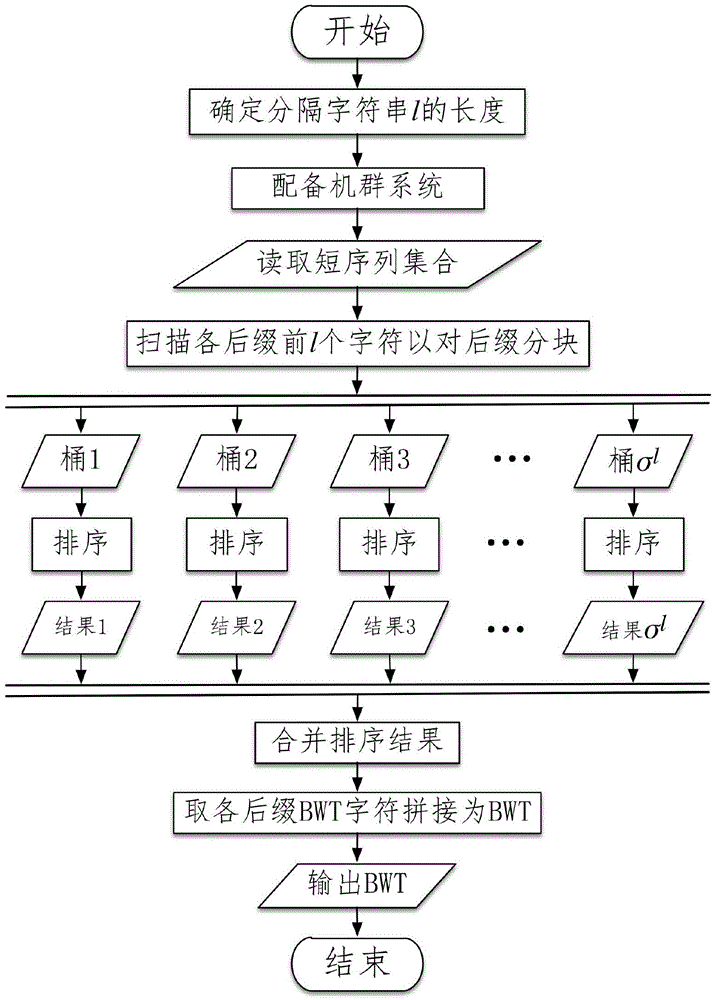
[0029] Attached below figure 1 , taking the construction of a BWT index of 1 billion DNA sequences with a length of 100 on a cluster with 64GB of memory at each node as an example (hereinafter referred to as this example), the present invention will be further described in detail. The alphabet of DNA sequence Σ={A, C, G, T}, the size is 4, that is, σ=4.
[0030] like figure 1 As shown, the novel BWT index parallel acceleration algorithm proposed by the present invention mainly includes 8 steps.
[0031] Step 1: Determine the length l of the delimited string used when the suffix is divided into blocks according to the sequence scale and the processor memory size, take For this example, m=10 9 , k=100, σ=4, M=64×2 30 , calculated to get
[0032] Step 2: Calculate σ l =4 4 =256, let’s equip a cluster system containing 256 processors (CPU), respectively numbered as p 1 ,p 2 ,...,p 256 .
[0033] Step 3: Open up σ in the cluster system memory l = 256 buckets, the l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com