DNA molecular chain displacement reaction based method for extracting CRNs for realizing combinational logic
A technology for strand displacement reaction and DNA molecules, applied in the field of DNA computing, can solve problems such as many reaction equations, high cost, and complexity, and achieve the effects of simple and convenient operation, reduced energy consumption, and simplified CRNs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
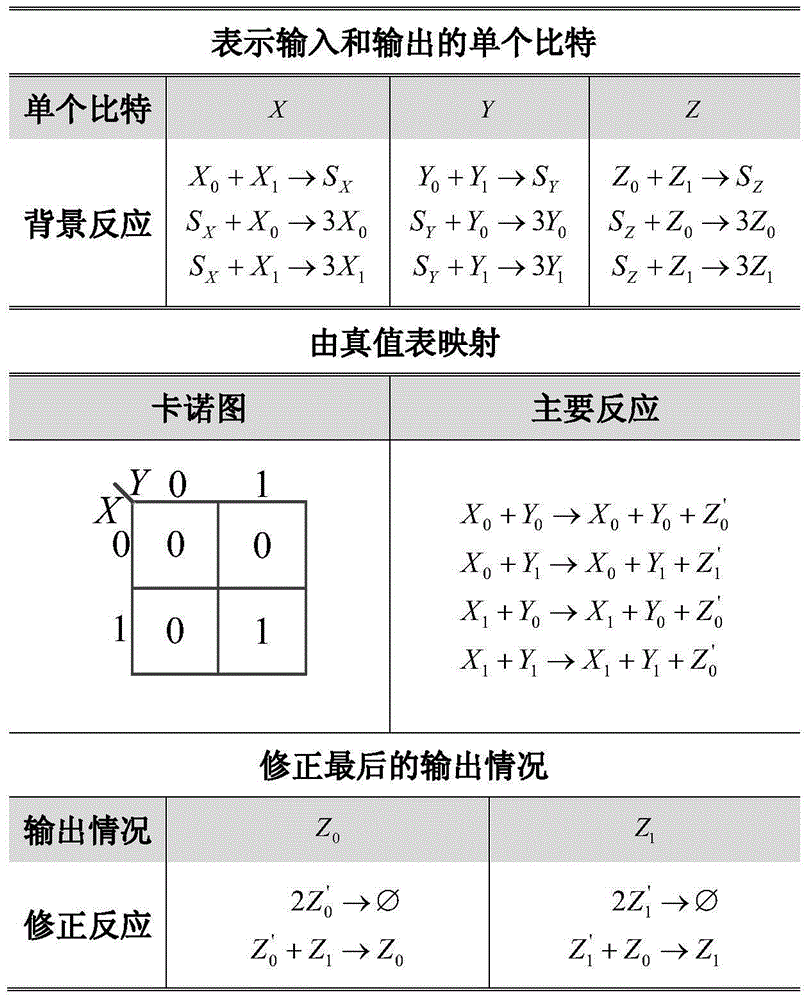
[0043] In the case of 2 inputs and 1 output, set the truth table, as shown in Table 2.
[0044] Table 2: Truth Table of Example 1
[0045] X Y Z 0 0 0 0 1 0 1 0 0 1 1 1
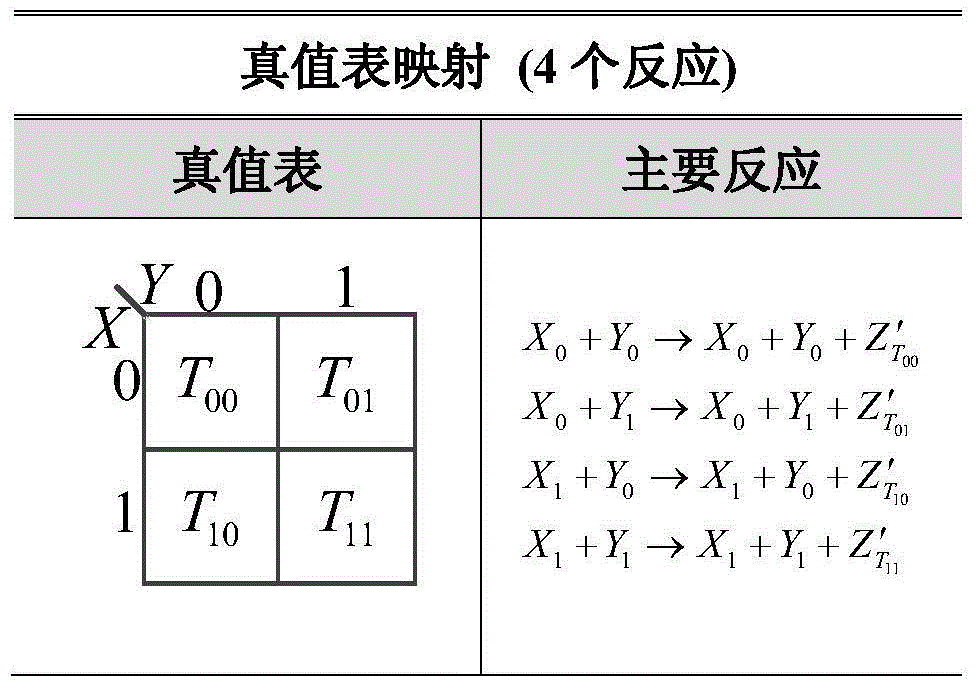
[0046] Such as figure 2 As shown, a set of CRNs containing 17 chemical reaction equations is obtained by combining the three reactions with the truth table and Karnaugh map.
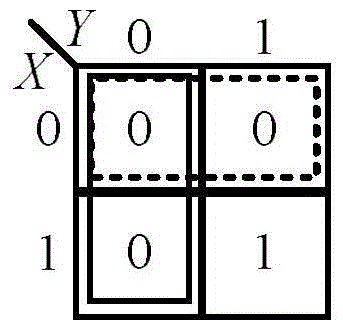
[0047] The CRNs composed of the above 17 chemical reaction equations were simplified by Karnaugh map simplification method. Such as image 3 As shown, according to the simplification method, only two Carnot circles of "0" cells can be circled, which are represented by dashed and solid lines respectively.
[0048] After simplification, the dotted box represents the input signal X 0 with Z 1 Generate Z 0 and x 0 , namely X 0 +Z 1 →X 0 +Z 0 , the solid line box represents the input signal Y 0 with Z 1 Generate Z 0 and Y 0 , namely Y 0 +Z 1 →Y 0 +Z 0 . Since all the "0" grids are ...
Embodiment 2
[0051] 3 inputs and 1 output, set the truth table, as shown in Table 3.
[0052] Table 3: Truth Table of Example 2
[0053] X Y O Z 0 0 0 0 0 0 1 1 0 1 0 1 0 1 1 0 1 0 0 0 1 0 1 1 1 1 0 1 1 1 1 0
[0054] Among them, the input signals are X, Y, O respectively, and the output signal is Z. If fully mapped according to the truth table, a total of 12 background responses, 8 main responses and 4 correction responses are required. Since there are no more than two reactants in a chemical reaction, it is necessary to downgrade 3 input 1 output to 2 input 1 output, so the intermediate reactant I is introduced 1 , I 2 , I 3 , I 4 . They represent the input signal corresponding to the simplification of each column of Carnot circles. Taking the first column as an example, it means the input signal Y 0 and O 0 Generate I 1 ; The second column represents the input signal Y 0 and O 1 Generate I 2 , and so on...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com