DNA sequencing method and next generation sequencing library
A sequencing library and sequencing technology, applied in the field of molecular biology, can solve the problems of directly distinguishing the target single-stranded DNA starting DNA template quantity requirement, the starting DNA template quantity requirement is low, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1D
[0049] Example 1DNA sample preparation
[0050] The sample to be prepared is viral single-stranded DNA, and the DNA is extracted using QIAampUltraSensVirusKit (QIAGEN). Proceed as follows:
[0051] 1) Use NL-DK1 cells in 5% fetal bovine serum to culture single-stranded DNA virus (Canine parvovirus canine parvovirus), take 1ml of the culture solution into a 2ml EP tube, add 0.8ml BufferAC, add 5.6μl carrierRNA, and mix;
[0052] 2) The mixed sample was left at room temperature for 10 minutes;
[0053] 3) Centrifuge at 1200×g for 3 minutes, discard the supernatant;
[0054] 4) Add 300 μl BufferAR preheated at 60°C and 20 μl proteinase K, and mix until the precipitate is completely suspended in the liquid;
[0055] 5) Incubate at 40°C for 10 minutes, and mix the samples every 5 minutes;
[0056] 6) Add 300μl BufferAB and mix well;
[0057] 7) Add the mixed liquid to the adsorption column, centrifuge at 3000-5000×g for 1 minute, discard the waste liquid, and transfer the adso...
Embodiment 2 2
[0062] Example 2 Construction and sequencing of next-generation sequencing library
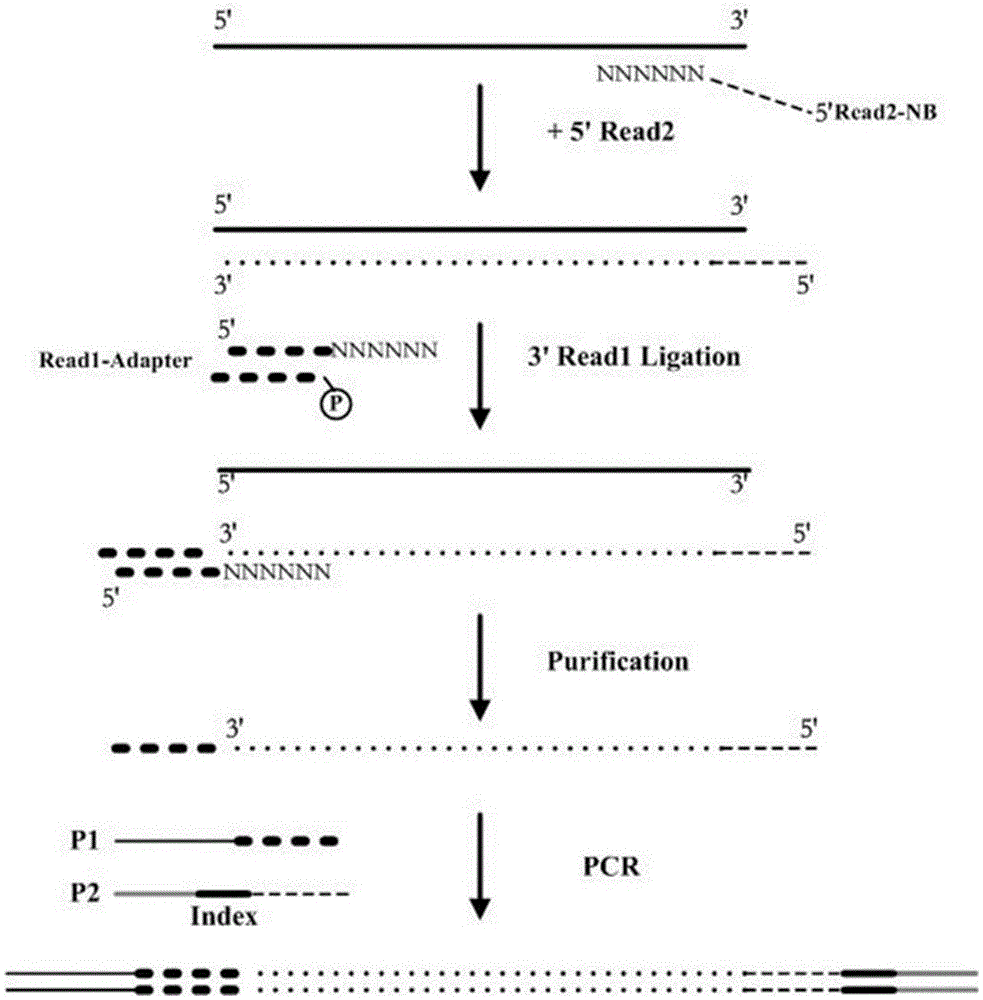
[0063] Rationale for Sequencing Methods figure 1 .
[0064] 1) Complementary extension:
[0065] Add the following reaction system to the reaction tube:
[0066]
[0067]
[0068] Carry out the following reaction procedure: 1-5min at 95°C; 10min at 25°C; pause at 37°C, and continue the reaction after adding the following reagents;
[0069] Add 0.5 μl of Klenow (NEB) to the reaction tube;
[0070] The following reaction procedure is carried out: 37°C for 30 minutes; 70°C for 10 minutes; and storage at 10°C to obtain a reaction product containing a complementary strand of viral single-stranded DNA linked with a specific sequence.
[0071] The above-mentioned DNA added to the reaction tube is the viral single-stranded DNA prepared in Example 1, and the above-mentioned Read2-NB-1 (SEQ ID NO: 4) is a random primer complementary to the viral single-stranded DNA, and the nucleotide sequence i...
Embodiment 3
[0109] Example 3 Analysis of sequencing results
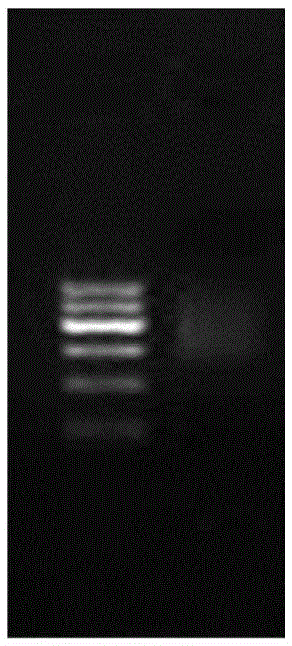
[0110] 1) The single-stranded DNA sequencing library constructed in Example 2 was sequenced on the Illumina sequencing platform, 251 cycles;
[0111] 2) Using the above method, the following results were obtained: a total of 7073 pairs of sequences were obtained, and after removing repeated sequences and low-quality sequences, a total of 6893 pairs of high-quality sequences (that is, obtained after removing repeated sequences and sequences whose quality did not reach Q20 sequence with unique features);
[0112] 3) The obtained high-quality sequences were assembled using the software SOAPdenovov2.04 (http: / / soap.genomics.org.cn / ), and a contig was obtained, the contigN50 was 5323 (genome size was 5323bp), and the sequence obtained by sequencing covered all Genome.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com