A kit for guiding the administration of tumor-targeted drugs
A tumor targeting and kit technology, applied in DNA/RNA fragments, measuring devices, instruments, etc., can solve problems such as differences in therapeutic effects, and achieve the effects of less denaturation and inactivation, sensitive detection, and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1H
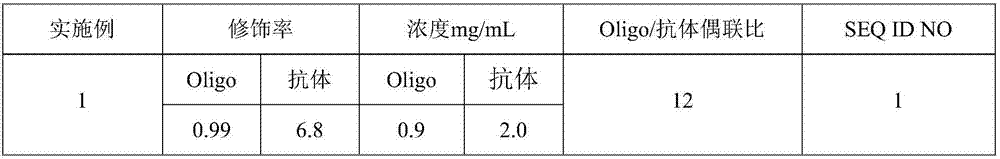
[0041] Embodiment 1 HER2-oligonucleotide probe
[0042] (1) Aldehyde modification of oligonucleotide probes
[0043] 150 nmol of Oligo1 (oligonucleotide) was prepared into a solution with 0.1 M pH 7.4 phosphate buffer. Weigh 1500nmol of SFB (N-succinimidyl 4-formylbenzoate), dissolve it with anhydrous DMF (N,N-dimethylformamide), react at room temperature for 2.5h, and purify through a column to obtain Oligo -FB (aldehyde modified oligonucleotide).
[0044] Detection of the concentration of Oligo-FB: detection of A by Nanodrop spectrophotometer 260 The calculated value of Oligo-FB is 2.6nmol / μL.
[0045] Detection of aldehyde group modification rate: Use quantitative 2-hydrazinopyridine-2-hydrochloride solution to detect the aldehyde group modification rate. Take the above-mentioned Oligo-FB and add it to the 2-hydrazinopyridine-2-hydrochloride solution, shake and mix well, and react at 37°C for 1 hour. The absorbance value at 360nm detected by Nanodrop is 6.3, and the modif...
Embodiment 2
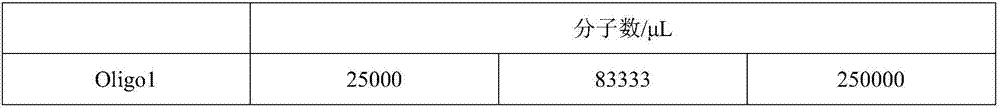
[0058] Example 2 QPCR amplification-specific detection of Oligo molecules
[0059] The Oligo1-13 molecules were diluted to three concentrations of 25000 molecules / μL, 83333 molecules / μL and 250000 molecules / μL. Then, taking Oligo1-3 as an example, Oligo1-3 was mixed at the same concentration to obtain mixed samples A, B, and C, as shown in Table 3.
[0060] Table 3 Example 2 sample formula
[0061]
[0062]
[0063] The above samples were configured with QPCR amplification solution according to Table 4 to obtain a QPCR amplification kit, and then Oligo1-3 was subjected to QPCR amplification according to the procedures shown in Table 5, and Oligo1-3 in mixed samples A, B, and C Each was subjected to QPCR amplification, using the extended Oligo1-3 as a template, and the Ct values of the amplification results are shown in Table 6. Among them, the extension primer is RT-P, its 3' end is complementary to the 3' end of the oligonucleotide, the 5' end can form a hairpin str...
Embodiment 3
[0078] Embodiment 3 makes standard curve
[0079] The same result can be obtained with Oligo1-13. In order to simplify the process, the following examples use Oligo1-4 as an example.
[0080] According to the steps of Example 1, Oligo2 was modified with SFB having a molar equivalent of 5 times to obtain Oligo-FB, and SANH with a molar equivalent of 10 times was used to modify EGFR with a hydrazine group to obtain EGFR-SANH, and the molar ratio was 10:1 EGFR-Oligo2 was obtained after reacting Oligo-FB with EGFR-SANH at room temperature for 16 hours.
[0081] According to the steps of Example 1, Oligo3 was modified with 20-fold molar equivalent of SFB to obtain Oligo-FB, and PD-L1 was hydrazino-modified with 50-fold molar equivalent of SANH to obtain PD-L1-SANH. The molar ratio PD-L1-Oligo3 was obtained after 8:1 Oligo-FB and PD-L1-SANH were reacted at room temperature for 24 hours.
[0082] According to the steps of Example 1, Oligo4 was modified with 10-fold molar equivalent...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com