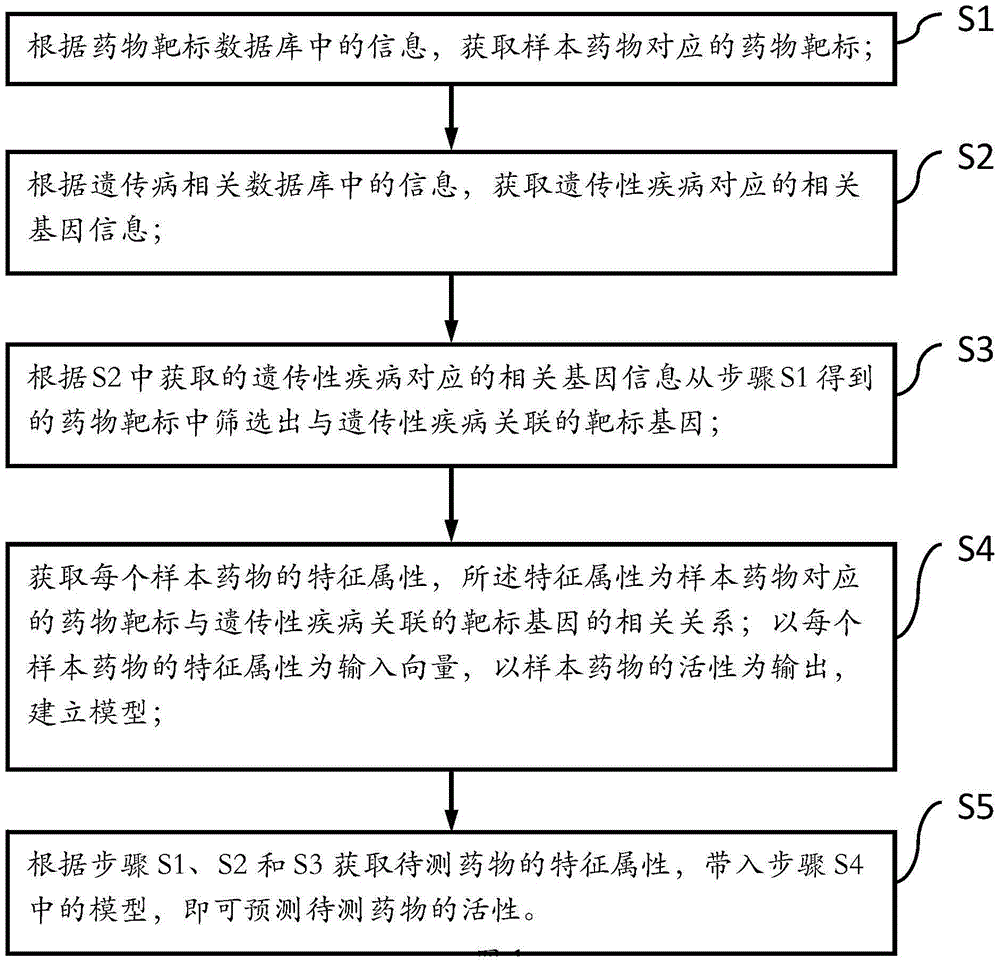
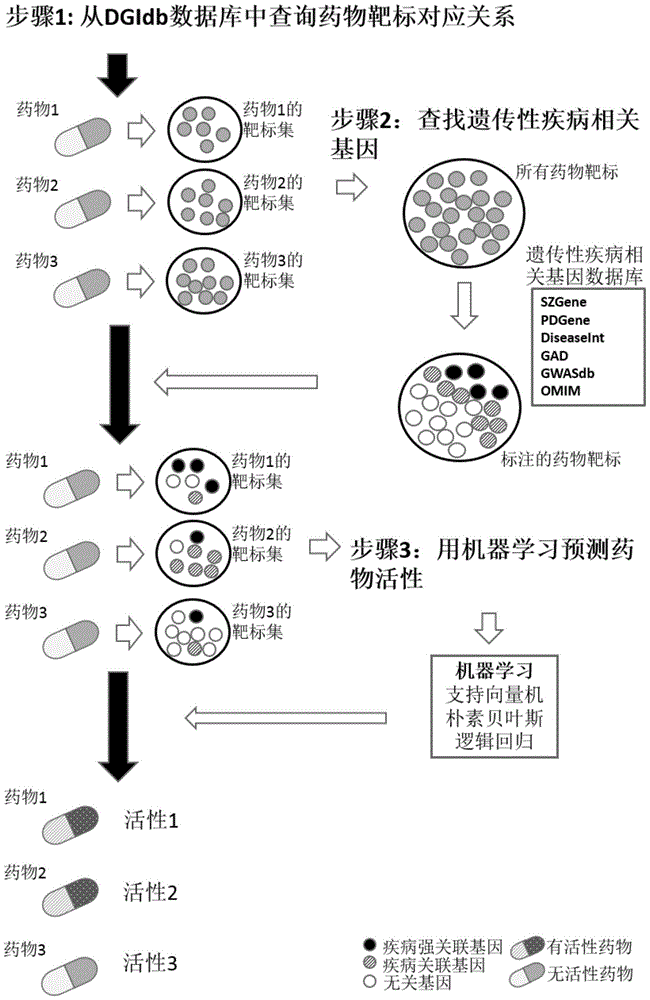
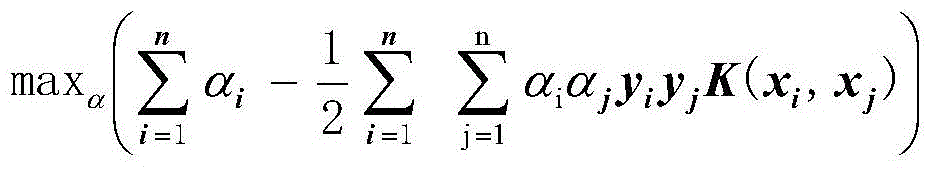Drug activity prediction method based on machine learning
A technology of drug activity and machine learning, applied in the fields of genomics, instrumentation, informatics, etc., can solve problems such as failure to obtain the expected return on investment, and achieve the effect of low cost, high efficiency, and improved efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Using the method of the present invention to predict the drug activity for the treatment of schizophrenia
[0065] 1. Collect human successfully marketed or researched drugs and their targets
[0066] Search the drug target database (including DGIdb: http: / / dgidb.genome.wustl.edu / ), and get a batch of drug targets corresponding to the drug. Taking DGIdb as the starting point, this experiment found a total of 2,271 targets with clear drug activity (the drug corresponds to the treatment of diseases), and 3,678 drugs corresponding to the above targets.
[0067] 2. Search for genes related to genetic information of schizophrenia
[0068] Genetic information-related genes of schizophrenia are composed of two parts of information. The first part is to find a total of 940 schizophrenia-related genes through the SZGene (http: / / www.szgene.org / ) database, and the second part is through the GWASdb ( http: / / jjwanglab.org / gwasdb ), GAD ( http: / / geneticassociationdb.nih.gov / ...
Embodiment 2
[0097] Using the method of the present invention to predict the drug activity for the treatment of Parkin's syndrome
[0098] 1. Collect human successfully marketed or researched drugs and their targets
[0099] Search the drug target database (including DGIdb: http: / / dgidb.genome.wustl.edu / ), and get a batch of drug targets corresponding to the drug. In this experiment, DGIdb was used as the starting point to find a total of 2,348 targets with clear drug activity (the drug corresponds to the treatment of diseases), and 3,678 drugs corresponding to the above targets.
[0100] 2. Find genes related to Parkin's syndrome genetic information
[0101] Genetic information related to Parkin's syndrome is composed of two parts of information. The first part is to find a total of 87 genes related to Parkin's syndrome through the PDGene (http: / / www.pdgene.org / ) database, and the second part is to use the GWASdb ( http: / / jjwanglab.org / gwasdb ), GAD ( http: / / geneticassociationdb.n...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com