Method for connecting DNA fragments through T4 bacteriophage DNA topoisomerase
A technology of topoisomerase and bacteriophage, applied in the field of biochemistry, can solve the problems of many steps, time-consuming, various cumbersome and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0014] A method utilizing T4 bacteriophage DNA topoisomerase I to connect DNA fragments, the specific steps are as follows:
[0015] Step 1, design primers: connect two DNA fragments with a length of 1.5kb, primers A, B and primers C, D each amplify a fragment with a length of 1.5kb, wherein primers A and D only contain sequences complementary to the template , primers B and C include linker DNA sequence, T4 phage DNA topoisomerase I recognition site DNA sequence and template paired DNA sequence from 5' to 3' end in turn, and the linker DNA sequence of primers 2 and 3 is reverse complementary. The source, size and primers A, B, C and D of DNA fragments are shown in Table 1
[0016] Table 1 DNA fragments and primers
[0017]
[0018] Bold letters indicate the linker sequence, and the underline indicates the T4 phage DNA topoisomerase I recognition site sequence.
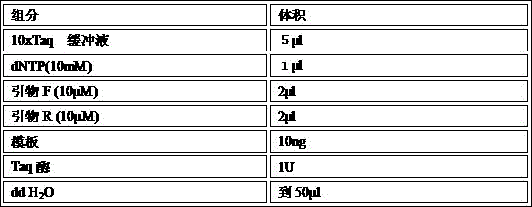
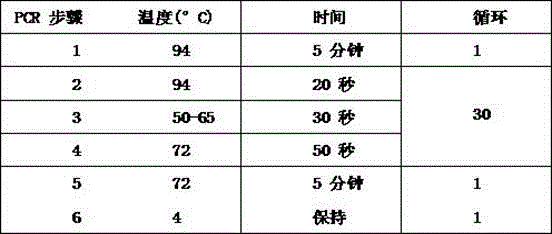
[0019] Step 2, fragment amplification: use the corresponding DNA fragments to be ligated as templates, and per...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com