Competitive compositions of nucleic acid molecules for enrichment of rare-allele-bearing species
A composition, nucleic acid technology, applied in combinatorial chemistry, organic chemistry, sugar derivatives, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0163] Experimental Results of Conditional Fluorescent Competitive Compositions
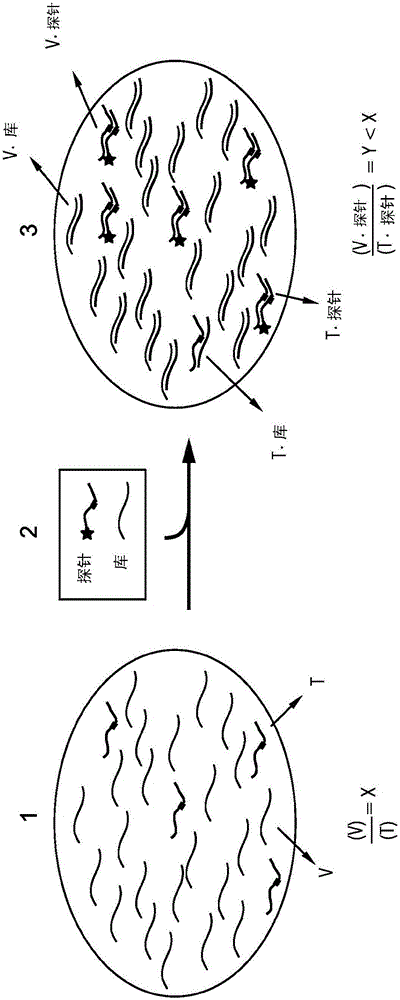
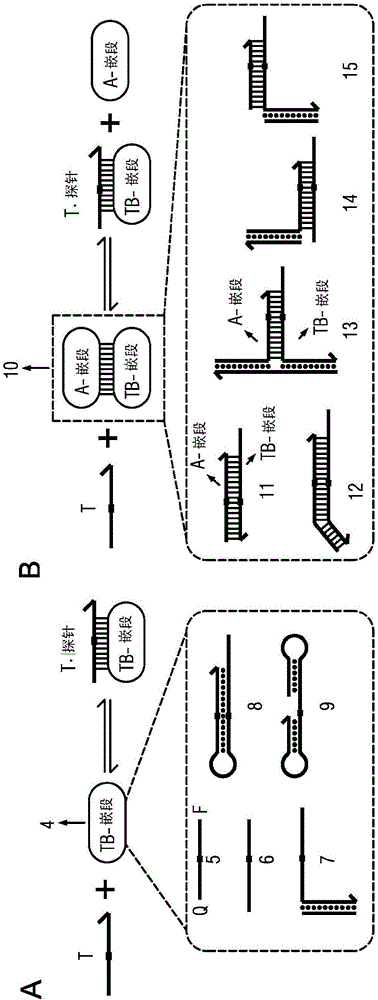
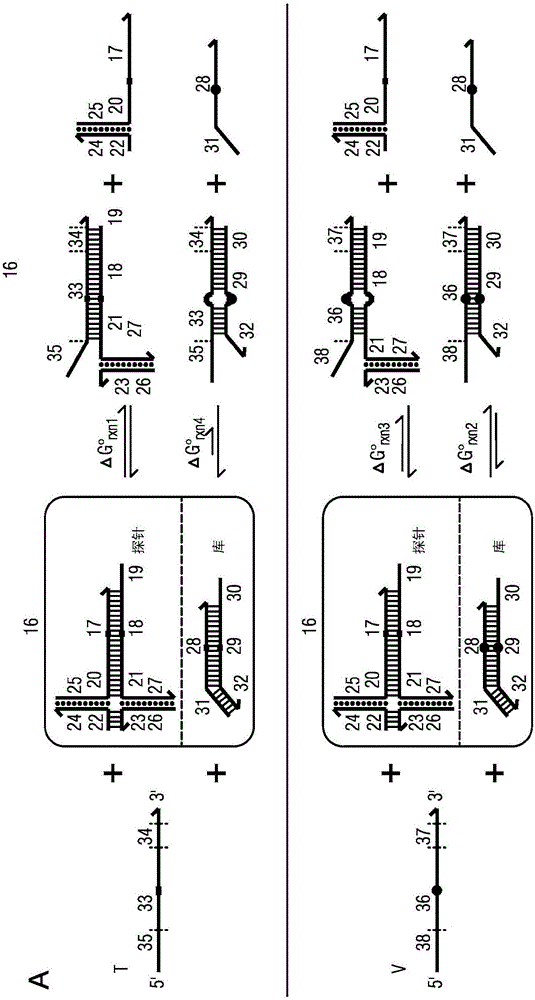
[0164] To experimentally verify the ability of the competing composition to enrich the target on the variants upon binding to the probe, experiments were designed and performed on conditionally fluorescent versions of the probe ( Figure 5 ). A probe is naturally dark until it hybridizes to a target or variant. By observing the fluorescent response to a sample of the variant with a small (loaded) amount of target, and the same amount of variant as in the absence of target, it can be deduced that enrichment is provided by the competing composition.
[0165] now refer to Figure 5 , 4 main products through image 3 The reactions described were formed; among these, 2 parameters involving the probe resulted in high fluorescence. The fluorescence generated by the target bound to the probe is denoted as f A , the fluorescence generated by the variant bound to the probe is denoted as f C , and the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com