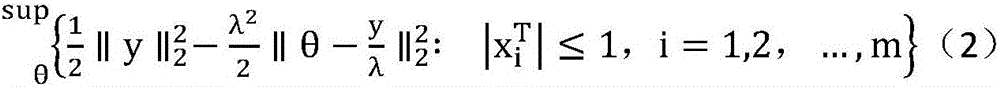Ligand molecule massive characteristic screening method in drug design
A ligand molecule and feature screening technology, which is applied in molecular design, calculation, chemical statistics, etc., can solve the problems of high time consumption and achieve the effect of increasing comprehensibility and improving learning efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018] The present invention will be further described in detail below in conjunction with the accompanying drawings.
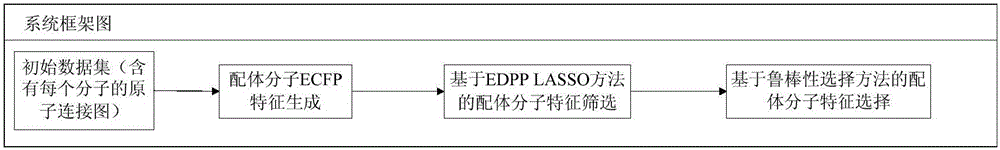
[0019] figure 1 It is a framework diagram of the system of the present invention. Based on the framework, the present invention provides a method for screening a large number of features of LASSO ligands based on the EDPP criterion. The specific implementation steps of the method include the following:
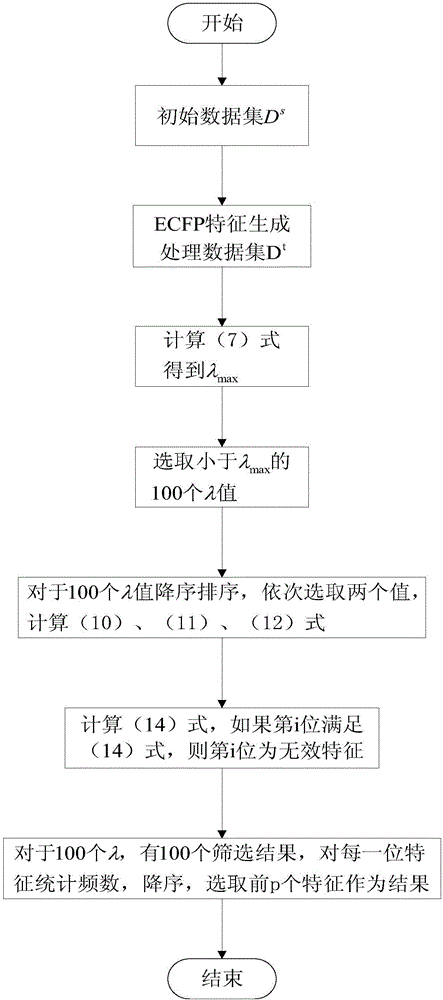
[0020] Step 1: Ligand molecule ECFP feature generation. Given an initial dataset in is the atomic connectivity graph of each molecule, Y i is the label for each sample. The initial data set is processed to obtain the ECFP characteristics describing the sample, that is, the data set D t ={(X i ,Y i )|X i ∈ R 1*m ,1≤i≤n}.
[0021] Step 2: Ligand molecular feature screening based on EDPP LASSO method. For data set D t , applying the EDPP criterion, for satisfying conditions (λ∈(0,λ 0 ]) of λ={λ i |0≤ii >λ i+1}, get the feature screening result o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com