Method for developing meconopsis SSR (simple sequence repeat) primers on basis of transcriptome sequences
A technology of transcriptome sequence and Meconopsis, applied in biochemical equipment and methods, determination/inspection of microorganisms, etc., can solve the problem of a small number of SSR primers, and achieve the effect of improving efficiency and increasing original data
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
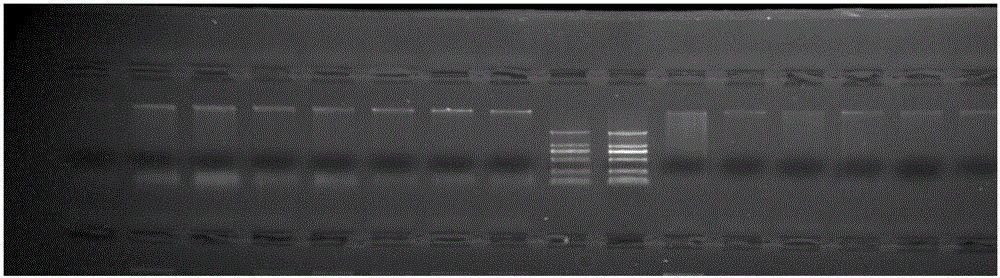
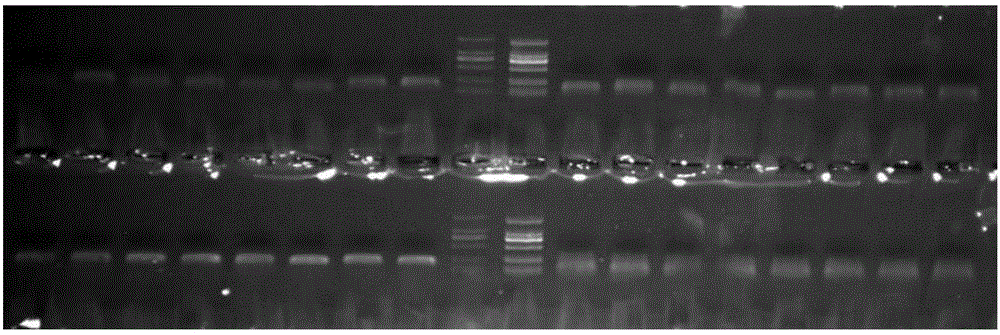
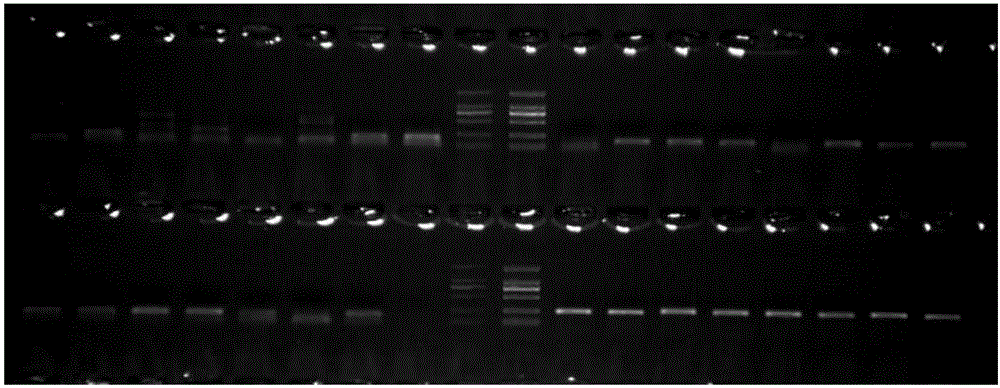
[0032] The method for developing Meconopsis SSR primers based on the transcriptome sequence in this embodiment, the specific steps are as follows:
[0033] (1) Construction of transcriptome library: Trizol was used to extract total RNA from Meconopsis petals, and the purified total RNA was subjected to mRNA isolation, fragmentation, first-strand cDNA synthesis, and second-strand cDNA synthesis according to the "TruSeq RNASample Preparation Guide" , using AMPureXP beads to purify cDNA; the purified double-stranded cDNA is then subjected to end repair, A-tailing and sequencing adapters, and then AMPureXP beads are used for fragment size selection; finally, a cDNA library is obtained by PCR enrichment; the built sequencing library is passed 0Fluorometer and Agilent2100 system passed the test, and completed paired-end sequencing on the Illumina HiSeq2000 sequencer; a total of 6,614 SSRs were detected and distributed in 17,311 Unigenes.
[0034] (2) After the sequencing is complet...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com